Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
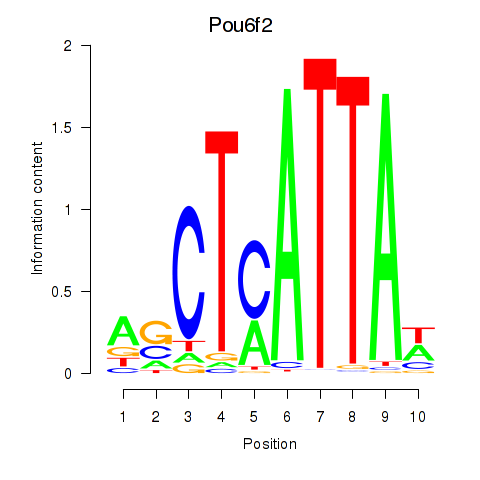
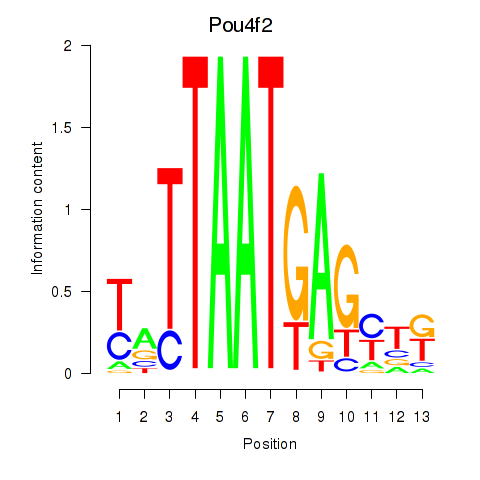
Results for Pou6f2_Pou4f2
Z-value: 0.85
Transcription factors associated with Pou6f2_Pou4f2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou6f2
|
ENSMUSG00000009734.19 | Pou6f2 |
|
Pou4f2
|
ENSMUSG00000031688.5 | Pou4f2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou6f2 | mm39_v1_chr13_-_18556626_18556626 | 0.68 | 7.5e-11 | Click! |
| Pou4f2 | mm39_v1_chr8_-_79163269_79163278 | 0.55 | 7.3e-07 | Click! |
Activity profile of Pou6f2_Pou4f2 motif
Sorted Z-values of Pou6f2_Pou4f2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou6f2_Pou4f2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_70491764 | 12.58 |
ENSMUST00000162144.2
ENSMUST00000162793.8 |
Phyhipl
|
phytanoyl-CoA hydroxylase interacting protein-like |
| chr12_-_112477536 | 9.29 |
ENSMUST00000066791.7
|
Tmem179
|
transmembrane protein 179 |
| chr1_+_153541412 | 7.94 |
ENSMUST00000111814.8
ENSMUST00000111810.2 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr4_-_110144676 | 7.88 |
ENSMUST00000106598.8
ENSMUST00000102723.11 ENSMUST00000153906.2 |
Elavl4
|
ELAV like RNA binding protein 4 |
| chr1_+_153541020 | 7.17 |
ENSMUST00000152114.8
ENSMUST00000111812.8 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr17_+_85335775 | 6.01 |
ENSMUST00000024944.9
|
Slc3a1
|
solute carrier family 3, member 1 |
| chr15_+_83676140 | 5.92 |
ENSMUST00000172115.8
ENSMUST00000172398.2 |
Mpped1
|
metallophosphoesterase domain containing 1 |
| chr1_+_172168764 | 5.91 |
ENSMUST00000056136.4
|
Kcnj10
|
potassium inwardly-rectifying channel, subfamily J, member 10 |
| chr11_-_42070517 | 5.61 |
ENSMUST00000206105.2
ENSMUST00000153147.3 |
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr13_-_56444118 | 5.18 |
ENSMUST00000224801.2
|
Cxcl14
|
chemokine (C-X-C motif) ligand 14 |
| chr1_+_153541339 | 4.64 |
ENSMUST00000147700.8
ENSMUST00000147482.8 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr2_-_92222979 | 4.55 |
ENSMUST00000111279.9
|
Mapk8ip1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr9_-_53882530 | 4.50 |
ENSMUST00000048409.14
|
Elmod1
|
ELMO/CED-12 domain containing 1 |
| chr14_-_9015639 | 4.47 |
ENSMUST00000112656.4
|
Synpr
|
synaptoporin |
| chr18_-_32082624 | 4.29 |
ENSMUST00000064016.6
|
Gpr17
|
G protein-coupled receptor 17 |
| chr6_-_138399896 | 4.13 |
ENSMUST00000161450.8
ENSMUST00000163024.8 ENSMUST00000162185.8 |
Lmo3
|
LIM domain only 3 |
| chr16_-_45544960 | 3.87 |
ENSMUST00000096057.5
|
Tagln3
|
transgelin 3 |
| chr10_+_75402090 | 3.84 |
ENSMUST00000129232.8
ENSMUST00000143792.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr12_+_52746158 | 3.78 |
ENSMUST00000095737.5
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr9_-_123507847 | 3.63 |
ENSMUST00000170591.2
ENSMUST00000171647.9 |
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr17_-_90395568 | 3.62 |
ENSMUST00000173222.2
|
Nrxn1
|
neurexin I |
| chr2_+_96148418 | 3.53 |
ENSMUST00000135431.8
ENSMUST00000162807.9 |
Lrrc4c
|
leucine rich repeat containing 4C |
| chr11_+_69217078 | 3.37 |
ENSMUST00000018614.3
|
Kcnab3
|
potassium voltage-gated channel, shaker-related subfamily, beta member 3 |
| chr8_-_68270870 | 3.36 |
ENSMUST00000059374.5
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr6_+_77219627 | 3.34 |
ENSMUST00000159616.2
|
Lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr17_+_17622934 | 3.24 |
ENSMUST00000115576.3
|
Lix1
|
limb and CNS expressed 1 |
| chr1_-_158183894 | 3.24 |
ENSMUST00000004133.11
|
Brinp2
|
bone morphogenic protein/retinoic acid inducible neural-specific 2 |
| chr6_+_77219698 | 3.18 |
ENSMUST00000161677.2
|
Lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr3_+_68479578 | 3.16 |
ENSMUST00000170788.9
|
Schip1
|
schwannomin interacting protein 1 |
| chr7_+_143729250 | 3.02 |
ENSMUST00000105900.9
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr2_-_79287095 | 2.93 |
ENSMUST00000041099.5
|
Neurod1
|
neurogenic differentiation 1 |
| chrX_-_58211440 | 2.85 |
ENSMUST00000119306.2
|
Fgf13
|
fibroblast growth factor 13 |
| chr16_-_96971905 | 2.75 |
ENSMUST00000056102.9
|
Dscam
|
DS cell adhesion molecule |
| chr14_-_9015757 | 2.72 |
ENSMUST00000153954.8
|
Synpr
|
synaptoporin |
| chr8_-_68270936 | 2.67 |
ENSMUST00000120071.8
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr9_-_99302205 | 2.66 |
ENSMUST00000123771.2
|
Mras
|
muscle and microspikes RAS |
| chr1_+_194302123 | 2.60 |
ENSMUST00000027952.12
|
Plxna2
|
plexin A2 |
| chr17_-_49871291 | 2.57 |
ENSMUST00000224595.2
ENSMUST00000057610.8 |
Daam2
|
dishevelled associated activator of morphogenesis 2 |
| chr12_+_38831093 | 2.56 |
ENSMUST00000161513.9
|
Etv1
|
ets variant 1 |
| chr13_-_78344492 | 2.56 |
ENSMUST00000125176.3
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr6_-_138404076 | 2.49 |
ENSMUST00000203435.3
|
Lmo3
|
LIM domain only 3 |
| chrX_+_113384008 | 2.43 |
ENSMUST00000113371.8
ENSMUST00000040504.12 |
Klhl4
|
kelch-like 4 |
| chr10_+_40225272 | 2.29 |
ENSMUST00000044672.11
ENSMUST00000095743.4 |
Cdk19
|
cyclin-dependent kinase 19 |
| chr10_+_79746690 | 2.29 |
ENSMUST00000181321.2
|
Gm26602
|
predicted gene, 26602 |
| chr6_-_138403732 | 2.28 |
ENSMUST00000162932.2
|
Lmo3
|
LIM domain only 3 |
| chr9_+_15150341 | 2.26 |
ENSMUST00000034413.8
|
Vstm5
|
V-set and transmembrane domain containing 5 |
| chr9_+_53678801 | 2.17 |
ENSMUST00000048670.10
|
Slc35f2
|
solute carrier family 35, member F2 |
| chr8_-_84184978 | 2.13 |
ENSMUST00000081506.11
|
Scoc
|
short coiled-coil protein |
| chr4_-_91288221 | 2.08 |
ENSMUST00000102799.10
|
Elavl2
|
ELAV like RNA binding protein 1 |
| chr9_+_32027335 | 2.07 |
ENSMUST00000174641.8
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr4_-_110148081 | 2.05 |
ENSMUST00000142722.2
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr12_+_38830812 | 2.04 |
ENSMUST00000160856.8
|
Etv1
|
ets variant 1 |
| chr6_+_29859372 | 2.04 |
ENSMUST00000115238.10
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr14_-_78866714 | 2.02 |
ENSMUST00000228362.2
ENSMUST00000227767.2 |
Dgkh
|
diacylglycerol kinase, eta |
| chr16_-_59452883 | 1.89 |
ENSMUST00000118438.8
|
Arl6
|
ADP-ribosylation factor-like 6 |
| chr5_+_29400981 | 1.85 |
ENSMUST00000160888.8
ENSMUST00000159272.8 ENSMUST00000001247.12 ENSMUST00000161398.8 ENSMUST00000160246.8 |
Rnf32
|
ring finger protein 32 |
| chr12_+_38830283 | 1.82 |
ENSMUST00000162563.8
ENSMUST00000161164.8 ENSMUST00000160996.8 |
Etv1
|
ets variant 1 |
| chr3_-_88363704 | 1.76 |
ENSMUST00000141471.2
ENSMUST00000123753.8 |
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr6_+_63232955 | 1.72 |
ENSMUST00000095852.5
|
Grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr4_+_150322151 | 1.68 |
ENSMUST00000141931.2
|
Eno1
|
enolase 1, alpha non-neuron |
| chr4_-_58499398 | 1.57 |
ENSMUST00000107570.2
|
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr1_-_9368721 | 1.55 |
ENSMUST00000132064.8
|
Sntg1
|
syntrophin, gamma 1 |
| chr9_-_60557076 | 1.54 |
ENSMUST00000053171.14
|
Lrrc49
|
leucine rich repeat containing 49 |
| chr5_+_117979899 | 1.54 |
ENSMUST00000142742.9
|
Nos1
|
nitric oxide synthase 1, neuronal |
| chr6_+_29859685 | 1.47 |
ENSMUST00000134438.2
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr6_+_106095726 | 1.47 |
ENSMUST00000113258.8
ENSMUST00000079416.6 |
Cntn4
|
contactin 4 |
| chr6_+_29859660 | 1.45 |
ENSMUST00000128927.9
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr19_-_18978463 | 1.43 |
ENSMUST00000040153.15
ENSMUST00000112828.8 |
Rorb
|
RAR-related orphan receptor beta |
| chr17_+_6025861 | 1.36 |
ENSMUST00000142409.8
ENSMUST00000061091.14 |
Synj2
|
synaptojanin 2 |
| chr19_-_55229668 | 1.34 |
ENSMUST00000069183.8
|
Gucy2g
|
guanylate cyclase 2g |
| chr3_+_76500857 | 1.33 |
ENSMUST00000162471.2
|
Fstl5
|
follistatin-like 5 |
| chr3_-_108133914 | 1.27 |
ENSMUST00000141387.4
|
Sypl2
|
synaptophysin-like 2 |
| chr13_+_110063364 | 1.22 |
ENSMUST00000117420.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr2_+_84564394 | 1.16 |
ENSMUST00000238573.2
ENSMUST00000090729.9 |
Ypel4
|
yippee like 4 |
| chr10_+_101517348 | 1.14 |
ENSMUST00000179929.8
ENSMUST00000219195.2 ENSMUST00000127504.9 |
Mgat4c
|
MGAT4 family, member C |
| chr4_-_116263183 | 1.13 |
ENSMUST00000123072.8
ENSMUST00000144281.2 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr17_+_6026015 | 1.11 |
ENSMUST00000115790.8
|
Synj2
|
synaptojanin 2 |
| chr3_-_79645101 | 1.10 |
ENSMUST00000078527.13
|
Rxfp1
|
relaxin/insulin-like family peptide receptor 1 |
| chr7_+_92729067 | 1.10 |
ENSMUST00000051179.12
|
Fam181b
|
family with sequence similarity 181, member B |
| chrX_+_9751861 | 1.09 |
ENSMUST00000067529.9
ENSMUST00000086165.4 |
Sytl5
|
synaptotagmin-like 5 |
| chr14_-_70043079 | 1.08 |
ENSMUST00000022665.4
|
Rhobtb2
|
Rho-related BTB domain containing 2 |
| chrX_+_16485937 | 1.02 |
ENSMUST00000026013.6
|
Maoa
|
monoamine oxidase A |
| chr7_-_115630282 | 1.00 |
ENSMUST00000206034.2
ENSMUST00000106612.8 |
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr10_+_18345706 | 0.99 |
ENSMUST00000162891.8
ENSMUST00000100054.4 |
Nhsl1
|
NHS-like 1 |
| chr7_+_51528788 | 0.97 |
ENSMUST00000107591.9
|
Gas2
|
growth arrest specific 2 |
| chr9_+_24194729 | 0.96 |
ENSMUST00000154644.2
|
Npsr1
|
neuropeptide S receptor 1 |
| chr19_-_5610628 | 0.96 |
ENSMUST00000025861.3
|
Ovol1
|
ovo like zinc finger 1 |
| chr9_+_113641615 | 0.93 |
ENSMUST00000111838.10
ENSMUST00000166734.10 ENSMUST00000214522.2 ENSMUST00000163895.3 |
Clasp2
|
CLIP associating protein 2 |
| chr2_+_71359000 | 0.90 |
ENSMUST00000126400.2
|
Dlx1
|
distal-less homeobox 1 |
| chr1_+_143653001 | 0.79 |
ENSMUST00000189936.7
ENSMUST00000018333.13 ENSMUST00000185493.7 |
Uchl5
|
ubiquitin carboxyl-terminal esterase L5 |
| chr3_+_55689921 | 0.79 |
ENSMUST00000075422.6
|
Mab21l1
|
mab-21-like 1 |
| chr4_+_150321659 | 0.73 |
ENSMUST00000133839.8
|
Eno1
|
enolase 1, alpha non-neuron |
| chr14_-_51134930 | 0.71 |
ENSMUST00000227271.2
|
Klhl33
|
kelch-like 33 |
| chr2_+_172994841 | 0.67 |
ENSMUST00000029017.6
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr1_-_143652711 | 0.59 |
ENSMUST00000159879.2
|
Ro60
|
Ro60, Y RNA binding protein |
| chr1_+_133109059 | 0.58 |
ENSMUST00000187285.7
|
Plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr4_+_117706390 | 0.57 |
ENSMUST00000132043.9
ENSMUST00000169990.8 |
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr4_+_117706559 | 0.56 |
ENSMUST00000163288.2
|
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr2_-_151510453 | 0.55 |
ENSMUST00000180195.8
ENSMUST00000096439.4 |
Rad21l
|
RAD21-like (S. pombe) |
| chr3_+_29568055 | 0.54 |
ENSMUST00000140288.2
|
Egfem1
|
EGF-like and EMI domain containing 1 |
| chr2_-_88559941 | 0.53 |
ENSMUST00000099815.2
|
Olfr1197
|
olfactory receptor 1197 |
| chr2_+_3115250 | 0.52 |
ENSMUST00000072955.12
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr8_-_3674993 | 0.50 |
ENSMUST00000142431.8
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr19_+_41921903 | 0.47 |
ENSMUST00000224258.2
ENSMUST00000026154.9 ENSMUST00000224896.2 |
Zdhhc16
|
zinc finger, DHHC domain containing 16 |
| chr14_-_52628228 | 0.45 |
ENSMUST00000078171.2
|
Olfr1511
|
olfactory receptor 1511 |
| chr11_-_100653754 | 0.45 |
ENSMUST00000107360.3
ENSMUST00000055083.4 |
Hcrt
|
hypocretin |
| chr7_-_10292412 | 0.45 |
ENSMUST00000236246.2
|
Vmn1r68
|
vomeronasal 1 receptor 68 |
| chr11_-_99213769 | 0.41 |
ENSMUST00000038004.3
|
Krt25
|
keratin 25 |
| chr10_+_101517556 | 0.39 |
ENSMUST00000156751.8
|
Mgat4c
|
MGAT4 family, member C |
| chr11_-_105346120 | 0.36 |
ENSMUST00000138977.8
|
Marchf10
|
membrane associated ring-CH-type finger 10 |
| chr12_-_81579614 | 0.33 |
ENSMUST00000169158.2
ENSMUST00000164431.2 ENSMUST00000163402.8 ENSMUST00000166664.2 ENSMUST00000164386.8 |
Synj2bp
Gm20498
|
synaptojanin 2 binding protein predicted gene 20498 |
| chr6_+_88701578 | 0.32 |
ENSMUST00000150180.4
ENSMUST00000163271.8 |
Mgll
|
monoglyceride lipase |
| chr19_+_13890894 | 0.31 |
ENSMUST00000216623.2
ENSMUST00000216835.2 |
Olfr1505
|
olfactory receptor 1505 |
| chr7_+_23400128 | 0.30 |
ENSMUST00000226233.2
ENSMUST00000227987.2 |
Vmn1r173
|
vomeronasal 1 receptor 173 |
| chr7_+_30193047 | 0.29 |
ENSMUST00000058280.13
ENSMUST00000133318.8 ENSMUST00000142575.8 ENSMUST00000131040.2 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chr6_+_134958681 | 0.27 |
ENSMUST00000167323.3
|
Apold1
|
apolipoprotein L domain containing 1 |
| chr11_-_120344299 | 0.24 |
ENSMUST00000026452.3
|
Pde6g
|
phosphodiesterase 6G, cGMP-specific, rod, gamma |
| chr10_+_73782857 | 0.23 |
ENSMUST00000191709.6
ENSMUST00000193739.6 ENSMUST00000195531.6 |
Pcdh15
|
protocadherin 15 |
| chr2_+_87576198 | 0.21 |
ENSMUST00000217572.2
|
Olfr1140
|
olfactory receptor 1140 |
| chr8_-_25215778 | 0.20 |
ENSMUST00000171438.8
ENSMUST00000171611.9 |
Adam3
|
a disintegrin and metallopeptidase domain 3 (cyritestin) |
| chr17_-_37399343 | 0.19 |
ENSMUST00000207101.2
ENSMUST00000217397.2 ENSMUST00000215195.2 ENSMUST00000216488.2 |
Olfr90
|
olfactory receptor 90 |
| chr17_+_38145802 | 0.19 |
ENSMUST00000050255.5
|
Olfr125
|
olfactory receptor 125 |
| chr2_-_17465410 | 0.18 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr8_-_3675024 | 0.17 |
ENSMUST00000133459.8
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr2_-_88157559 | 0.14 |
ENSMUST00000214207.2
|
Olfr1175
|
olfactory receptor 1175 |
| chr2_-_86109346 | 0.14 |
ENSMUST00000217294.2
ENSMUST00000217245.2 ENSMUST00000216432.2 |
Olfr1051
|
olfactory receptor 1051 |
| chr4_+_102446883 | 0.14 |
ENSMUST00000097949.11
ENSMUST00000106901.2 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr17_-_3608056 | 0.14 |
ENSMUST00000041003.8
|
Tfb1m
|
transcription factor B1, mitochondrial |
| chr4_+_118516149 | 0.12 |
ENSMUST00000213189.3
|
Olfr62
|
olfactory receptor 62 |
| chr8_-_5155347 | 0.12 |
ENSMUST00000023835.3
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr4_+_5724305 | 0.11 |
ENSMUST00000108380.2
|
Fam110b
|
family with sequence similarity 110, member B |
| chr18_+_57601541 | 0.10 |
ENSMUST00000091892.4
ENSMUST00000209782.2 |
Ctxn3
|
cortexin 3 |
| chr10_-_129107354 | 0.08 |
ENSMUST00000204573.3
|
Olfr777
|
olfactory receptor 777 |
| chr10_+_99099084 | 0.05 |
ENSMUST00000020118.5
ENSMUST00000220291.2 |
Dusp6
|
dual specificity phosphatase 6 |
| chr13_+_24023386 | 0.05 |
ENSMUST00000039721.14
|
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr19_+_13674632 | 0.05 |
ENSMUST00000214007.2
ENSMUST00000216377.2 ENSMUST00000215493.2 ENSMUST00000216366.2 ENSMUST00000216622.2 |
Olfr1491
|
olfactory receptor 1491 |
| chr11_+_73851643 | 0.04 |
ENSMUST00000213134.2
ENSMUST00000216291.2 |
Olfr397
|
olfactory receptor 397 |
| chr7_-_106531426 | 0.04 |
ENSMUST00000215468.2
|
Olfr709-ps1
|
olfactory receptor 709, pseudogene 1 |
| chr7_-_28038129 | 0.03 |
ENSMUST00000209141.2
ENSMUST00000003527.10 |
Supt5
|
suppressor of Ty 5, DSIF elongation factor subunit |
| chr4_+_54947976 | 0.03 |
ENSMUST00000098070.10
|
Zfp462
|
zinc finger protein 462 |
| chr18_-_15197138 | 0.02 |
ENSMUST00000234864.2
|
Kctd1
|
potassium channel tetramerisation domain containing 1 |
| chr7_+_23239157 | 0.02 |
ENSMUST00000235361.2
|
Vmn1r168
|
vomeronasal 1 receptor 168 |
| chrX_+_152020744 | 0.02 |
ENSMUST00000112574.9
|
Klf8
|
Kruppel-like factor 8 |
| chr11_+_115225557 | 0.02 |
ENSMUST00000106543.8
ENSMUST00000019006.5 |
Otop3
|
otopetrin 3 |
| chr18_-_88945571 | 0.01 |
ENSMUST00000147313.2
|
Socs6
|
suppressor of cytokine signaling 6 |
| chr4_+_56740070 | 0.01 |
ENSMUST00000181745.2
|
Gm26657
|
predicted gene, 26657 |
| chr1_+_11063678 | 0.01 |
ENSMUST00000027056.12
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr13_-_24098981 | 0.01 |
ENSMUST00000110407.4
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr4_+_119397710 | 0.01 |
ENSMUST00000160219.2
|
Foxj3
|
forkhead box J3 |
| chr13_-_24098951 | 0.01 |
ENSMUST00000021769.16
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr13_+_24023428 | 0.01 |
ENSMUST00000091698.12
ENSMUST00000110422.3 ENSMUST00000166467.9 |
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr8_-_22422724 | 0.01 |
ENSMUST00000062586.3
|
Defb15
|
defensin beta 15 |
| chr7_-_119122681 | 0.00 |
ENSMUST00000033267.4
|
Pdilt
|
protein disulfide isomerase-like, testis expressed |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 5.9 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 1.3 | 19.8 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 1.3 | 5.2 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.9 | 2.7 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.9 | 3.6 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.9 | 2.6 | GO:0021934 | hindbrain tangential cell migration(GO:0021934) |
| 0.7 | 4.6 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.6 | 3.8 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.6 | 3.6 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.6 | 2.9 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.6 | 5.0 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.5 | 3.8 | GO:1901748 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.4 | 2.9 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.4 | 1.6 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.4 | 1.5 | GO:1901204 | regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.4 | 1.9 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.4 | 1.1 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.4 | 1.1 | GO:0060618 | nipple development(GO:0060618) |
| 0.3 | 6.4 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.3 | 1.0 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.3 | 0.9 | GO:1902871 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.3 | 6.5 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.2 | 1.0 | GO:2000292 | negative regulation of eating behavior(GO:1903999) regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.2 | 4.3 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.2 | 0.7 | GO:0006114 | glycerol biosynthetic process(GO:0006114) positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 0.2 | 2.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.2 | 6.0 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 1.7 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.1 | 1.0 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.1 | 3.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 2.8 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 1.4 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 3.2 | GO:0001553 | luteinization(GO:0001553) |
| 0.1 | 1.0 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 | 1.4 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 2.6 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 1.0 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 0.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 3.0 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 0.9 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 2.0 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 8.4 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 9.9 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.1 | 3.1 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 1.0 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 | 0.6 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 0.2 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.1 | 0.4 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 1.8 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.1 | 0.6 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 3.5 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 3.2 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.8 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 6.0 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 1.3 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 2.6 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 1.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 4.7 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 2.3 | GO:0050729 | positive regulation of inflammatory response(GO:0050729) |
| 0.0 | 2.4 | GO:0071229 | cellular response to acid chemical(GO:0071229) |
| 0.0 | 0.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.9 | 3.8 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.6 | 9.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.4 | 19.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.3 | 0.9 | GO:1904511 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.3 | 8.5 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 5.9 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.2 | 5.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 3.0 | GO:0005883 | neurofilament(GO:0005883) |
| 0.2 | 3.6 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 2.5 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.2 | 2.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.6 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 1.5 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 6.0 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.8 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 1.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 6.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 9.8 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 2.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.7 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 1.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.6 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 5.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 3.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.9 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.7 | 5.0 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.7 | 5.6 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.4 | 3.6 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.3 | 21.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.3 | 3.8 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.3 | 1.5 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.3 | 1.4 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.3 | 3.6 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.3 | 4.6 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 9.9 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.2 | 0.7 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.2 | 3.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 2.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 2.5 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.2 | 2.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 2.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 6.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 5.2 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 2.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.0 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 1.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 1.0 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 10.1 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 2.7 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 1.7 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.1 | 3.0 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 1.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 2.9 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 2.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 2.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.8 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 3.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 5.6 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 1.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.6 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 3.7 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 2.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 2.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 2.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 2.7 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 3.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 6.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.8 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.2 | 5.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 4.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.7 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 5.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 21.3 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 6.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.0 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 2.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 3.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 3.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 3.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 2.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.8 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 4.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 2.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 2.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |