Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
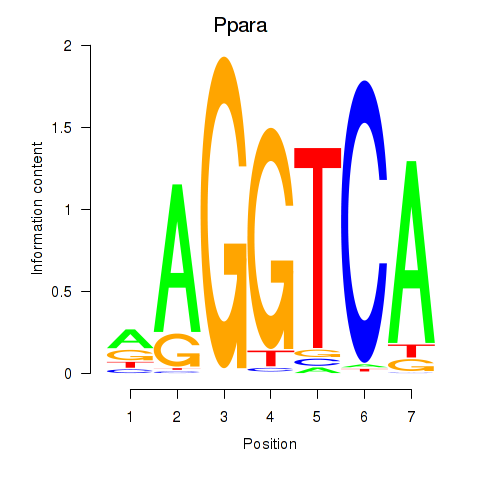
Results for Ppara
Z-value: 2.25
Transcription factors associated with Ppara
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ppara
|
ENSMUSG00000022383.14 | Ppara |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ppara | mm39_v1_chr15_+_85620308_85620332 | -0.17 | 1.5e-01 | Click! |
Activity profile of Ppara motif
Sorted Z-values of Ppara motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Ppara
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.2 | 48.9 | GO:0021933 | radial glia guided migration of cerebellar granule cell(GO:0021933) negative regulation of cerebellar granule cell precursor proliferation(GO:0021941) |
| 10.4 | 31.3 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 6.4 | 19.3 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 5.9 | 29.6 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 5.6 | 16.8 | GO:0086097 | actin filament uncapping(GO:0051695) phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 5.5 | 21.9 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 4.9 | 14.7 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 4.4 | 13.1 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 4.1 | 12.2 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 3.9 | 15.7 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 3.7 | 11.1 | GO:1900247 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 3.7 | 11.0 | GO:2000331 | regulation of terminal button organization(GO:2000331) |
| 3.6 | 14.5 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 3.2 | 9.7 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 3.0 | 9.0 | GO:0060466 | activation of meiosis involved in egg activation(GO:0060466) |
| 2.9 | 8.8 | GO:0061193 | taste bud development(GO:0061193) |
| 2.9 | 5.8 | GO:1901079 | positive regulation of relaxation of muscle(GO:1901079) |
| 2.8 | 11.3 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 2.8 | 25.3 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 2.7 | 10.9 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 2.5 | 15.1 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 2.5 | 12.3 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 2.4 | 12.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 2.4 | 9.7 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 2.3 | 4.5 | GO:0033292 | T-tubule organization(GO:0033292) |
| 2.2 | 11.1 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 2.1 | 8.6 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 2.1 | 10.7 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 2.1 | 6.4 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 2.1 | 10.6 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 2.1 | 10.3 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 2.1 | 47.4 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 2.0 | 10.2 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 2.0 | 6.0 | GO:0033058 | directional locomotion(GO:0033058) |
| 1.9 | 5.8 | GO:0097115 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 1.9 | 7.4 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 1.8 | 11.0 | GO:0046103 | inosine biosynthetic process(GO:0046103) |
| 1.8 | 5.4 | GO:1903116 | positive regulation of actin filament-based movement(GO:1903116) |
| 1.8 | 7.2 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 1.8 | 17.6 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 1.7 | 17.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 1.7 | 5.1 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 1.7 | 10.2 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 1.7 | 1.7 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 1.7 | 11.7 | GO:0035989 | tendon development(GO:0035989) |
| 1.6 | 4.9 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 1.6 | 32.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 1.6 | 25.5 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) |
| 1.6 | 9.6 | GO:0036394 | amylase secretion(GO:0036394) |
| 1.6 | 4.7 | GO:0042323 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 1.6 | 7.8 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 1.5 | 6.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 1.5 | 4.6 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 1.5 | 7.5 | GO:1902163 | negative regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902163) |
| 1.5 | 7.5 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 1.4 | 10.1 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 1.4 | 2.7 | GO:1901204 | regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 1.4 | 4.1 | GO:1903632 | positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) |
| 1.3 | 6.7 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 1.3 | 9.1 | GO:0048840 | otolith development(GO:0048840) |
| 1.3 | 9.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 1.3 | 6.4 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 1.2 | 3.7 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 1.2 | 6.2 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 1.2 | 11.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 1.2 | 4.8 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) |
| 1.2 | 28.0 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 1.2 | 12.7 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 1.1 | 5.7 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 1.1 | 4.6 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 1.1 | 4.3 | GO:0015744 | succinate transport(GO:0015744) |
| 1.1 | 7.5 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 1.1 | 4.2 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 1.0 | 4.2 | GO:2000984 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 1.0 | 7.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.0 | 1.0 | GO:1903181 | regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 1.0 | 10.9 | GO:0033227 | dsRNA transport(GO:0033227) |
| 1.0 | 4.9 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 1.0 | 2.9 | GO:0046166 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 1.0 | 3.9 | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 1.0 | 3.9 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 1.0 | 5.8 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 1.0 | 2.9 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 1.0 | 28.8 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 1.0 | 2.9 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 1.0 | 8.6 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.9 | 5.6 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.9 | 3.6 | GO:1902081 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.9 | 2.6 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.9 | 9.6 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.9 | 5.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.9 | 7.7 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.9 | 7.7 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.8 | 10.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.8 | 2.5 | GO:0007225 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.8 | 6.8 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) |
| 0.8 | 8.4 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.8 | 4.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.8 | 4.8 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.8 | 2.4 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.8 | 3.2 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.8 | 18.7 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.8 | 2.3 | GO:2000485 | asparagine transport(GO:0006867) regulation of glutamine transport(GO:2000485) positive regulation of glutamine transport(GO:2000487) |
| 0.8 | 2.3 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.8 | 3.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.8 | 6.2 | GO:0060313 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.8 | 2.3 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.8 | 2.3 | GO:0048352 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) positive regulation of cardiac ventricle development(GO:1904414) |
| 0.8 | 25.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.8 | 4.5 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.7 | 3.0 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.7 | 33.1 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.7 | 25.0 | GO:1905145 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.7 | 4.4 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.7 | 2.9 | GO:0021905 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.7 | 7.9 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) |
| 0.7 | 2.1 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.7 | 3.6 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.7 | 17.8 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.7 | 2.8 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.7 | 16.0 | GO:0007614 | short-term memory(GO:0007614) |
| 0.7 | 9.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.7 | 4.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.7 | 4.7 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.7 | 3.4 | GO:1904253 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.7 | 2.0 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.7 | 4.0 | GO:0015676 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.7 | 9.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.7 | 4.6 | GO:0015824 | proline transport(GO:0015824) |
| 0.6 | 13.6 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.6 | 3.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.6 | 4.4 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.6 | 9.4 | GO:0099624 | atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 0.6 | 10.6 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.6 | 5.6 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.6 | 4.3 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.6 | 9.6 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.6 | 1.2 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.6 | 3.6 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.6 | 9.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.6 | 1.8 | GO:0015827 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) positive regulation of gap junction assembly(GO:1903598) |
| 0.6 | 2.3 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.6 | 2.3 | GO:0032364 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) oxygen homeostasis(GO:0032364) gas homeostasis(GO:0033483) |
| 0.6 | 12.0 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.6 | 5.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.6 | 3.4 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.6 | 2.2 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.6 | 2.2 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.6 | 4.4 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.5 | 20.1 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.5 | 13.0 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.5 | 9.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.5 | 3.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.5 | 8.8 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.5 | 1.0 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.5 | 6.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.5 | 2.5 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.5 | 2.0 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.5 | 6.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.5 | 1.5 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.5 | 8.7 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.5 | 1.9 | GO:0000105 | histidine biosynthetic process(GO:0000105) |
| 0.5 | 2.4 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.5 | 6.6 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.5 | 23.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.5 | 1.4 | GO:0021629 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.5 | 4.7 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.5 | 4.2 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.5 | 5.9 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.4 | 2.7 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.4 | 13.4 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.4 | 1.3 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) malonyl-CoA metabolic process(GO:2001293) |
| 0.4 | 32.6 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.4 | 6.7 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.4 | 2.7 | GO:0060686 | esophagus smooth muscle contraction(GO:0014846) negative regulation of prostatic bud formation(GO:0060686) |
| 0.4 | 21.3 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.4 | 7.5 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.4 | 1.8 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.4 | 9.2 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.4 | 3.9 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.4 | 4.4 | GO:0086036 | regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 0.4 | 26.5 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.4 | 2.1 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.4 | 1.7 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.4 | 5.3 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.4 | 17.6 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.4 | 4.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.4 | 7.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.4 | 7.2 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.4 | 3.2 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.4 | 3.2 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.4 | 1.6 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.4 | 1.9 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.4 | 1.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.4 | 5.7 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.4 | 6.7 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.4 | 4.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.4 | 1.5 | GO:0032468 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.4 | 6.6 | GO:0003299 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.4 | 5.4 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.4 | 12.5 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.4 | 9.7 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.4 | 6.3 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.3 | 4.7 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.3 | 7.0 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.3 | 5.6 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.3 | 0.9 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.3 | 5.0 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.3 | 0.9 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.3 | 12.5 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.3 | 1.8 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.3 | 2.4 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.3 | 4.1 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.3 | 9.9 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.3 | 0.9 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.3 | 3.4 | GO:0048671 | regulation of collateral sprouting(GO:0048670) negative regulation of collateral sprouting(GO:0048671) |
| 0.3 | 3.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.3 | 11.8 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.3 | 10.7 | GO:0050807 | regulation of synapse organization(GO:0050807) |
| 0.3 | 1.7 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.3 | 23.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.3 | 6.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.3 | 17.4 | GO:1903078 | positive regulation of protein localization to plasma membrane(GO:1903078) |
| 0.3 | 6.5 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.3 | 2.4 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.3 | 5.1 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.3 | 3.5 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.3 | 21.9 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.3 | 1.6 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) regulation of NADP metabolic process(GO:1902031) |
| 0.3 | 1.3 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.3 | 1.0 | GO:0009305 | protein biotinylation(GO:0009305) response to biotin(GO:0070781) histone biotinylation(GO:0071110) |
| 0.3 | 10.4 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.3 | 8.1 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.3 | 3.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.3 | 5.3 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.2 | 10.7 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.2 | 13.0 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.2 | 2.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.2 | 4.8 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.2 | 3.8 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 2.9 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.2 | 2.7 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.2 | 4.8 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.2 | 1.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.2 | 37.2 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.2 | 4.6 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.2 | 5.0 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.2 | 0.8 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.2 | 2.5 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.2 | 2.3 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.2 | 2.1 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.2 | 0.8 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.2 | 6.1 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.2 | 1.4 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.2 | 10.7 | GO:1901099 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.2 | 4.6 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.2 | 1.6 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.2 | 2.6 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.2 | 9.1 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.2 | 1.8 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.2 | 0.8 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.2 | 10.5 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.2 | 1.0 | GO:0009946 | proximal/distal axis specification(GO:0009946) |
| 0.2 | 15.0 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.2 | 3.1 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.2 | 2.6 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.2 | 1.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 1.7 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.2 | 1.8 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.2 | 12.8 | GO:0050766 | positive regulation of phagocytosis(GO:0050766) |
| 0.2 | 6.1 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.2 | 8.8 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.2 | 5.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.2 | 0.9 | GO:0070295 | glycerol transport(GO:0015793) renal water absorption(GO:0070295) |
| 0.2 | 12.7 | GO:0031960 | response to corticosteroid(GO:0031960) response to glucocorticoid(GO:0051384) |
| 0.2 | 3.6 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.2 | 2.7 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.2 | 4.2 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.2 | 19.7 | GO:0006165 | nucleoside diphosphate phosphorylation(GO:0006165) |
| 0.2 | 0.3 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.2 | 6.5 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.2 | 0.8 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.2 | 2.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.2 | 1.4 | GO:0060613 | fat pad development(GO:0060613) |
| 0.2 | 1.4 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.2 | 1.4 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.2 | 9.0 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.2 | 2.8 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.2 | 10.5 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.2 | 4.8 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.2 | 5.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 7.8 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.1 | 1.5 | GO:0000423 | macromitophagy(GO:0000423) response to mitochondrial depolarisation(GO:0098780) |
| 0.1 | 9.6 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.1 | 4.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 8.6 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 1.0 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 8.8 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.1 | 51.6 | GO:0048812 | neuron projection morphogenesis(GO:0048812) |
| 0.1 | 1.4 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.1 | 0.4 | GO:0048822 | enucleate erythrocyte development(GO:0048822) |
| 0.1 | 1.4 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 3.5 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.1 | 1.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.8 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.6 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 3.6 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.1 | 0.7 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.8 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.4 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 0.6 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.5 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 2.6 | GO:0060004 | reflex(GO:0060004) |
| 0.1 | 0.8 | GO:0000022 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 1.1 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.1 | 0.5 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) peptidyl-histidine modification(GO:0018202) |
| 0.1 | 2.2 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.1 | 3.0 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.1 | 6.7 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.1 | 2.8 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 0.9 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 1.1 | GO:1904871 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.1 | 0.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.7 | GO:0042228 | interleukin-8 biosynthetic process(GO:0042228) regulation of interleukin-6 biosynthetic process(GO:0045408) regulation of interleukin-8 biosynthetic process(GO:0045414) |
| 0.1 | 3.2 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.1 | 0.7 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.1 | 1.2 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.1 | 1.5 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 3.1 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 2.7 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 2.3 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.8 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.1 | 0.2 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 0.2 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.1 | 0.6 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 2.6 | GO:0008361 | regulation of cell size(GO:0008361) |
| 0.0 | 2.6 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 2.9 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 6.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 2.4 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 2.0 | GO:0014033 | neural crest cell differentiation(GO:0014033) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 1.0 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.7 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 1.2 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 1.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.3 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 1.4 | GO:0051058 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.6 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 1.2 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.6 | GO:0050773 | regulation of dendrite development(GO:0050773) |
| 0.0 | 0.6 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.5 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 2.2 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 2.5 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.1 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.8 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 2.0 | GO:0030534 | adult behavior(GO:0030534) |
| 0.0 | 1.5 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 1.0 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.8 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.0 | 0.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 1.0 | GO:0018196 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.4 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.0 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 1.3 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 14.7 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 3.4 | 10.2 | GO:0098855 | HCN channel complex(GO:0098855) |
| 3.4 | 10.1 | GO:0090537 | CERF complex(GO:0090537) |
| 3.2 | 32.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 2.8 | 35.9 | GO:0097427 | microtubule bundle(GO:0097427) |
| 2.5 | 12.5 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 2.5 | 12.3 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 2.2 | 11.1 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 2.1 | 6.4 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 2.0 | 56.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 1.9 | 7.6 | GO:0072534 | perineuronal net(GO:0072534) |
| 1.8 | 5.4 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 1.6 | 7.9 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 1.5 | 10.6 | GO:0097444 | spine apparatus(GO:0097444) |
| 1.5 | 4.5 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 1.5 | 17.8 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 1.5 | 8.8 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 1.3 | 18.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 1.2 | 9.7 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 1.2 | 5.8 | GO:1990130 | Iml1 complex(GO:1990130) |
| 1.1 | 3.4 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 1.1 | 4.4 | GO:0014802 | terminal cisterna(GO:0014802) |
| 1.1 | 12.0 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 1.1 | 3.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 1.0 | 10.5 | GO:0070552 | BRISC complex(GO:0070552) |
| 1.0 | 33.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 1.0 | 5.8 | GO:0042583 | chromaffin granule(GO:0042583) |
| 1.0 | 12.4 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.9 | 5.7 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.9 | 6.6 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.9 | 5.6 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.8 | 37.9 | GO:0031430 | M band(GO:0031430) |
| 0.8 | 9.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.8 | 67.2 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.8 | 8.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.8 | 17.0 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.8 | 8.8 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.8 | 2.4 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.8 | 9.5 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.8 | 11.0 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.8 | 2.3 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.8 | 6.9 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.8 | 9.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.8 | 2.3 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.7 | 35.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.7 | 18.0 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.7 | 5.6 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.7 | 20.0 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.7 | 33.8 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.7 | 4.0 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.6 | 5.1 | GO:0001652 | granular component(GO:0001652) |
| 0.6 | 2.5 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.6 | 9.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.6 | 1.8 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.6 | 1.7 | GO:1990879 | CST complex(GO:1990879) |
| 0.5 | 2.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.5 | 15.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.5 | 6.0 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.5 | 2.6 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.5 | 54.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.5 | 4.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.5 | 26.4 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.5 | 7.9 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.5 | 3.4 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.5 | 2.9 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.5 | 4.7 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.4 | 3.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.4 | 37.6 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.4 | 2.6 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.4 | 7.4 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.4 | 13.0 | GO:0031672 | A band(GO:0031672) |
| 0.4 | 2.1 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.4 | 7.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.4 | 1.7 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.4 | 4.1 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.4 | 5.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.4 | 6.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.4 | 3.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.4 | 1.5 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.4 | 1.9 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.4 | 4.4 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.4 | 5.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.3 | 0.7 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.3 | 32.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.3 | 3.0 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.3 | 16.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.3 | 5.7 | GO:0036038 | MKS complex(GO:0036038) |
| 0.3 | 3.7 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.3 | 2.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.3 | 17.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 5.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.3 | 42.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.3 | 7.8 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.3 | 5.8 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.3 | 3.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.3 | 4.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.3 | 1.1 | GO:0005606 | laminin-1 complex(GO:0005606) laminin-3 complex(GO:0005608) |
| 0.3 | 2.9 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.3 | 10.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 11.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.2 | 55.6 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.2 | 53.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 1.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.2 | 21.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 51.0 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.2 | 0.7 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.2 | 4.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 2.8 | GO:0043679 | axon terminus(GO:0043679) |
| 0.2 | 3.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 1.6 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 1.4 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.2 | 1.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.2 | 1.4 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.2 | 1.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 2.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 4.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 18.6 | GO:0005901 | caveola(GO:0005901) |
| 0.2 | 1.6 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.2 | 19.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.2 | 1.7 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 2.0 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 11.3 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 2.9 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 11.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 2.0 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 1.2 | GO:0098794 | postsynapse(GO:0098794) |
| 0.1 | 2.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 38.3 | GO:0030425 | dendrite(GO:0030425) |
| 0.1 | 2.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 7.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 5.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 2.0 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 1.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 1.0 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 10.7 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 7.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 0.4 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 23.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 7.6 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 2.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 2.2 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 3.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 4.7 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 1.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 5.5 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 8.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 1.9 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 1.0 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 1.1 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 1.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 6.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 12.2 | GO:0045202 | synapse(GO:0045202) |
| 0.1 | 0.7 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 10.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 3.1 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 6.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 3.1 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 3.6 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 5.2 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 1.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.8 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.2 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 1.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.0 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 17.4 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 5.7 | 17.2 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 4.9 | 14.7 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 3.4 | 16.8 | GO:0051373 | FATZ binding(GO:0051373) |
| 3.3 | 10.0 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 3.0 | 18.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 2.9 | 8.8 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 2.8 | 31.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 2.7 | 32.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 2.7 | 10.6 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 2.5 | 10.1 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 2.2 | 17.6 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 2.2 | 21.9 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 2.1 | 6.4 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 2.1 | 41.1 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 1.9 | 18.9 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 1.9 | 7.4 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 1.8 | 10.9 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 1.8 | 9.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 1.8 | 14.5 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 1.8 | 18.1 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 1.8 | 16.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 1.8 | 1.8 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 1.8 | 56.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 1.7 | 10.2 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 1.6 | 4.9 | GO:0070279 | vitamin B6 binding(GO:0070279) |
| 1.6 | 6.2 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 1.5 | 7.3 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 1.4 | 4.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 1.4 | 4.3 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 1.4 | 7.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.3 | 7.9 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 1.3 | 9.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 1.3 | 18.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 1.3 | 5.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 1.3 | 15.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 1.3 | 8.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.3 | 8.8 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 1.2 | 7.5 | GO:0055100 | adiponectin binding(GO:0055100) |
| 1.2 | 13.5 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 1.2 | 3.7 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 1.2 | 4.6 | GO:0000010 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 1.2 | 8.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 1.1 | 9.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 1.1 | 7.9 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 1.1 | 16.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 1.1 | 4.4 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 1.1 | 15.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 1.1 | 7.5 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 1.1 | 3.2 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 1.0 | 14.5 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 1.0 | 21.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 1.0 | 2.9 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 1.0 | 2.9 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.9 | 2.8 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.9 | 10.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.9 | 14.1 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.9 | 7.9 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.9 | 7.0 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.9 | 25.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.8 | 3.4 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.8 | 6.7 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.8 | 5.0 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.8 | 34.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.8 | 3.2 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.8 | 3.2 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.8 | 2.3 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.8 | 2.3 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.8 | 15.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.7 | 3.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.7 | 3.6 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.7 | 7.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.7 | 7.9 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.7 | 2.9 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.7 | 4.9 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.7 | 5.6 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.7 | 11.1 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.7 | 6.7 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.6 | 36.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.6 | 8.0 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.6 | 12.7 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.6 | 15.4 | GO:0004629 | phospholipase C activity(GO:0004629) |
| 0.6 | 2.3 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) L-asparagine transmembrane transporter activity(GO:0015182) alanine transmembrane transporter activity(GO:0022858) |
| 0.6 | 2.3 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.6 | 1.7 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.6 | 5.0 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.6 | 3.9 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.5 | 1.6 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.5 | 14.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.5 | 14.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.5 | 2.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.5 | 3.0 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.5 | 2.0 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.5 | 12.7 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.5 | 5.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.5 | 15.9 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.5 | 2.4 | GO:0052795 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.5 | 3.8 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.5 | 1.4 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.5 | 4.7 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.5 | 20.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.5 | 12.8 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.5 | 3.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.4 | 2.7 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.4 | 1.3 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.4 | 2.2 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.4 | 1.8 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.4 | 4.4 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.4 | 1.8 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.4 | 35.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.4 | 2.2 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.4 | 2.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.4 | 6.0 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.4 | 17.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.4 | 3.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.4 | 23.8 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.4 | 42.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.4 | 9.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.4 | 5.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.4 | 34.3 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.4 | 4.2 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.4 | 15.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.4 | 2.7 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.4 | 3.4 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.4 | 2.6 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.4 | 5.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.4 | 22.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.4 | 18.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.4 | 3.0 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.4 | 1.5 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.4 | 8.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.4 | 4.3 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.3 | 11.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.3 | 1.0 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.3 | 2.0 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.3 | 4.7 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.3 | 1.0 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.3 | 2.3 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.3 | 21.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.3 | 3.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.3 | 1.3 | GO:0038100 | nodal binding(GO:0038100) |
| 0.3 | 2.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.3 | 16.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.3 | 1.9 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.3 | 11.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.3 | 1.9 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.3 | 8.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.3 | 5.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.3 | 2.9 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.3 | 0.9 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.3 | 1.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.3 | 13.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.3 | 3.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.3 | 5.9 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.3 | 9.5 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.3 | 7.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.3 | 1.4 | GO:0002046 | opsin binding(GO:0002046) |
| 0.3 | 2.5 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.3 | 1.9 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.3 | 0.8 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.3 | 1.3 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.3 | 2.4 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.3 | 2.4 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.3 | 8.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.3 | 1.0 | GO:0018271 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.3 | 2.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 2.2 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.2 | 15.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.2 | 1.7 | GO:0031419 | hemoglobin binding(GO:0030492) cobalamin binding(GO:0031419) |
| 0.2 | 1.7 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.2 | 5.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 1.6 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.2 | 1.4 | GO:1990269 | RNA polymerase II C-terminal domain phosphoserine binding(GO:1990269) |
| 0.2 | 1.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 6.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 1.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 10.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 1.4 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.2 | 2.7 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.2 | 21.5 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.2 | 3.7 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.2 | 6.3 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.2 | 9.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 5.0 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 8.0 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 10.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 0.9 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.2 | 1.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.2 | 3.0 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 1.0 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.2 | 0.8 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.2 | 1.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 4.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 24.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.2 | 34.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.2 | 0.6 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.2 | 0.8 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.2 | 1.4 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 5.1 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 2.4 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.1 | 3.8 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 0.5 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 3.1 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.5 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 2.1 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 2.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 9.6 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.1 | 4.2 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 2.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 2.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.5 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 6.0 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 7.9 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 9.7 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 10.8 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.1 | 1.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.5 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.6 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 28.9 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 3.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 6.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 23.6 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 7.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 1.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 1.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 2.0 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 2.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 2.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 4.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.2 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.1 | 3.2 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 1.8 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 0.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 2.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 0.6 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 5.3 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 0.8 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 5.3 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.2 | GO:0031370 | eukaryotic initiation factor 4G binding(GO:0031370) |
| 0.1 | 19.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 1.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.6 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.1 | 5.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 0.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.8 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 6.2 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 3.3 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 1.0 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 1.9 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 2.0 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 6.7 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 1.2 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 3.1 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 4.7 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 21.8 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 1.6 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 1.9 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.3 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 1.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.5 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 4.5 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 1.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 18.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.5 | 25.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.4 | 21.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.4 | 4.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.4 | 36.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.3 | 7.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.3 | 17.0 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.3 | 7.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.3 | 7.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.3 | 16.7 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.3 | 9.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.3 | 8.1 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.2 | 0.9 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.2 | 6.6 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.2 | 14.1 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.2 | 16.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 9.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 2.9 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 0.8 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 6.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 4.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 7.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 3.4 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 7.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 2.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 3.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 10.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 4.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 3.0 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 0.6 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 3.0 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 1.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 1.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 2.1 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 1.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 12.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 2.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 2.0 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 8.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.5 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 56.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 1.7 | 23.6 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 1.5 | 19.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 1.3 | 42.4 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 1.2 | 26.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.9 | 31.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.9 | 18.6 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.8 | 12.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.7 | 16.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.7 | 14.9 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.7 | 7.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.7 | 7.9 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.7 | 9.8 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.6 | 5.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.6 | 8.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.6 | 12.8 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.6 | 4.9 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.6 | 36.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.5 | 17.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.5 | 11.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.5 | 25.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.5 | 9.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.5 | 16.7 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.5 | 17.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.5 | 6.0 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.4 | 14.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.4 | 4.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.4 | 20.7 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.4 | 4.9 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.4 | 18.1 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.4 | 10.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.4 | 19.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.4 | 7.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.3 | 15.1 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.3 | 19.1 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.3 | 5.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.3 | 16.1 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.3 | 19.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 21.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.3 | 4.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.3 | 4.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.3 | 8.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 3.0 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 2.5 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.2 | 3.1 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.2 | 7.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.2 | 3.8 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.2 | 4.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 9.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.2 | 2.5 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.2 | 9.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.2 | 4.8 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 3.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 4.8 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 2.9 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 12.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 4.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 3.5 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 1.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 5.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 2.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 2.0 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 4.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 2.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 8.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 3.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 5.7 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 2.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 2.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.8 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 1.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 1.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 0.9 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.1 | 2.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 12.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 0.9 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 3.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 0.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.6 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.1 | 0.9 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 0.4 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 1.1 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 1.1 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.0 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 4.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.0 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.9 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.1 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 0.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 4.8 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 1.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 1.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.7 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.2 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 4.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |