Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Prdm1
Z-value: 1.59
Transcription factors associated with Prdm1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Prdm1
|
ENSMUSG00000038151.14 | Prdm1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Prdm1 | mm39_v1_chr10_-_44334683_44334710 | 0.10 | 3.9e-01 | Click! |
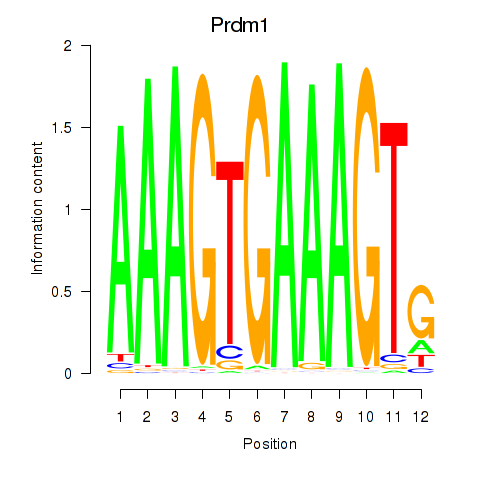
Activity profile of Prdm1 motif
Sorted Z-values of Prdm1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Prdm1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_114355789 | 22.71 |
ENSMUST00000103486.2
|
Ighv6-3
|
immunoglobulin heavy variable 6-3 |
| chr17_+_37504783 | 11.25 |
ENSMUST00000038844.7
|
Ubd
|
ubiquitin D |
| chr6_+_67586695 | 11.02 |
ENSMUST00000103303.3
|
Igkv1-135
|
immunoglobulin kappa variable 1-135 |
| chr6_+_40619913 | 10.96 |
ENSMUST00000238599.2
|
Mgam
|
maltase-glucoamylase |
| chr5_-_105130522 | 9.85 |
ENSMUST00000031239.13
|
Abcg3
|
ATP binding cassette subfamily G member 3 |
| chr5_+_31078775 | 9.45 |
ENSMUST00000201621.4
|
Khk
|
ketohexokinase |
| chr9_-_44714263 | 9.28 |
ENSMUST00000044694.8
|
Ttc36
|
tetratricopeptide repeat domain 36 |
| chr12_-_115109539 | 8.96 |
ENSMUST00000192554.6
ENSMUST00000103522.3 |
Ighv1-52
|
immunoglobulin heavy variable 1-52 |
| chr5_+_31078911 | 7.75 |
ENSMUST00000201571.4
|
Khk
|
ketohexokinase |
| chr13_-_3968157 | 7.74 |
ENSMUST00000223258.2
ENSMUST00000091853.12 |
Net1
|
neuroepithelial cell transforming gene 1 |
| chr9_+_5308828 | 7.17 |
ENSMUST00000162846.8
ENSMUST00000027012.14 |
Casp4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr3_+_106389732 | 7.13 |
ENSMUST00000029508.11
|
Dennd2d
|
DENN/MADD domain containing 2D |
| chr1_-_190915441 | 7.11 |
ENSMUST00000027941.14
|
Atf3
|
activating transcription factor 3 |
| chr13_+_74787952 | 7.03 |
ENSMUST00000221822.2
ENSMUST00000221526.2 |
Erap1
|
endoplasmic reticulum aminopeptidase 1 |
| chr16_+_10307589 | 6.97 |
ENSMUST00000230892.2
ENSMUST00000230146.2 ENSMUST00000230392.2 |
Ciita
|
class II transactivator |
| chr11_+_69855584 | 6.75 |
ENSMUST00000108597.8
ENSMUST00000060651.6 ENSMUST00000108596.8 |
Cldn7
|
claudin 7 |
| chr17_-_34406193 | 6.72 |
ENSMUST00000173831.3
|
Psmb9
|
proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2) |
| chr1_-_85247864 | 6.71 |
ENSMUST00000159582.8
ENSMUST00000161267.8 |
C130026I21Rik
|
RIKEN cDNA C130026I21 gene |
| chr12_-_115587215 | 6.57 |
ENSMUST00000199933.5
ENSMUST00000103539.3 |
Ighv1-69
|
immunoglobulin heavy variable 1-69 |
| chr2_+_43445333 | 6.53 |
ENSMUST00000028223.9
ENSMUST00000112826.8 |
Kynu
|
kynureninase |
| chr11_+_48967411 | 6.48 |
ENSMUST00000109202.3
|
Ifi47
|
interferon gamma inducible protein 47 |
| chr8_-_11528615 | 6.48 |
ENSMUST00000033900.7
|
Rab20
|
RAB20, member RAS oncogene family |
| chr14_+_40827108 | 6.38 |
ENSMUST00000224514.2
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr16_-_35691914 | 6.31 |
ENSMUST00000042665.9
|
Parp14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr6_+_117145496 | 6.21 |
ENSMUST00000112866.8
ENSMUST00000112871.8 ENSMUST00000073043.5 |
Cxcl12
|
chemokine (C-X-C motif) ligand 12 |
| chr11_-_77784922 | 6.18 |
ENSMUST00000017597.5
|
Pipox
|
pipecolic acid oxidase |
| chr6_-_55152002 | 6.13 |
ENSMUST00000003569.6
|
Inmt
|
indolethylamine N-methyltransferase |
| chr7_-_101234800 | 6.11 |
ENSMUST00000209526.2
|
Art2b
|
ADP-ribosyltransferase 2b |
| chr8_+_95161006 | 6.01 |
ENSMUST00000211816.2
|
Nlrc5
|
NLR family, CARD domain containing 5 |
| chr12_+_111383864 | 6.01 |
ENSMUST00000220537.2
ENSMUST00000223050.2 ENSMUST00000072646.8 ENSMUST00000223431.2 ENSMUST00000221144.2 ENSMUST00000222437.2 |
Exoc3l4
|
exocyst complex component 3-like 4 |
| chr15_+_77613239 | 6.00 |
ENSMUST00000230979.2
ENSMUST00000109775.4 |
Apol9b
|
apolipoprotein L 9b |
| chr6_+_41523664 | 5.98 |
ENSMUST00000103299.3
|
Trbc2
|
T cell receptor beta, constant 2 |
| chr6_-_48549594 | 5.92 |
ENSMUST00000009425.7
ENSMUST00000204267.3 ENSMUST00000204930.3 ENSMUST00000204182.2 |
Rarres2
|
retinoic acid receptor responder (tazarotene induced) 2 |
| chr14_+_40826970 | 5.76 |
ENSMUST00000225720.2
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr4_-_42773987 | 5.69 |
ENSMUST00000095114.5
|
Ccl21a
|
chemokine (C-C motif) ligand 21A (serine) |
| chr10_+_28544356 | 5.69 |
ENSMUST00000060409.13
ENSMUST00000056097.11 ENSMUST00000105516.9 |
Themis
|
thymocyte selection associated |
| chr17_-_32643067 | 5.62 |
ENSMUST00000237130.2
|
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr1_+_85528392 | 5.56 |
ENSMUST00000080204.11
|
Sp140
|
Sp140 nuclear body protein |
| chr10_-_75633362 | 5.52 |
ENSMUST00000120177.8
|
Gstt1
|
glutathione S-transferase, theta 1 |
| chr14_+_40827317 | 5.50 |
ENSMUST00000047286.7
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr2_+_85850977 | 5.44 |
ENSMUST00000213774.2
ENSMUST00000214546.2 ENSMUST00000215682.2 |
Olfr1033
|
olfactory receptor 1033 |
| chr11_+_120421496 | 5.40 |
ENSMUST00000026119.8
|
Gcgr
|
glucagon receptor |
| chr8_-_45786226 | 5.38 |
ENSMUST00000095328.6
|
Cyp4v3
|
cytochrome P450, family 4, subfamily v, polypeptide 3 |
| chr14_+_56813060 | 5.36 |
ENSMUST00000161553.2
|
Parp4
|
poly (ADP-ribose) polymerase family, member 4 |
| chr3_+_142326363 | 5.15 |
ENSMUST00000165774.8
|
Gbp2
|
guanylate binding protein 2 |
| chr1_+_139429430 | 5.10 |
ENSMUST00000027615.7
|
F13b
|
coagulation factor XIII, beta subunit |
| chr6_+_39358036 | 5.07 |
ENSMUST00000031986.5
|
Rab19
|
RAB19, member RAS oncogene family |
| chr6_+_71350411 | 5.03 |
ENSMUST00000066747.14
|
Cd8a
|
CD8 antigen, alpha chain |
| chr5_-_105387395 | 4.98 |
ENSMUST00000065588.7
|
Gbp10
|
guanylate-binding protein 10 |
| chr11_-_120618052 | 4.87 |
ENSMUST00000106148.10
ENSMUST00000026144.5 |
Dcxr
|
dicarbonyl L-xylulose reductase |
| chr7_+_43057611 | 4.86 |
ENSMUST00000005592.7
|
Siglecg
|
sialic acid binding Ig-like lectin G |
| chr17_-_36501112 | 4.85 |
ENSMUST00000025312.13
ENSMUST00000102675.10 |
H2-T3
|
histocompatibility 2, T region locus 3 |
| chr10_-_75353792 | 4.85 |
ENSMUST00000118936.8
|
Gucd1
|
guanylyl cyclase domain containing 1 |
| chr3_+_122688721 | 4.77 |
ENSMUST00000023820.6
|
Fabp2
|
fatty acid binding protein 2, intestinal |
| chr5_+_115061293 | 4.75 |
ENSMUST00000031540.11
ENSMUST00000112143.4 |
Oasl1
|
2'-5' oligoadenylate synthetase-like 1 |
| chr10_-_75353690 | 4.70 |
ENSMUST00000123505.2
ENSMUST00000147269.8 |
Gucd1
|
guanylyl cyclase domain containing 1 |
| chr13_+_24023428 | 4.61 |
ENSMUST00000091698.12
ENSMUST00000110422.3 ENSMUST00000166467.9 |
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr18_+_89215575 | 4.59 |
ENSMUST00000097496.4
ENSMUST00000236452.2 |
Cd226
|
CD226 antigen |
| chr11_-_99045894 | 4.53 |
ENSMUST00000103134.4
|
Ccr7
|
chemokine (C-C motif) receptor 7 |
| chr11_-_5900019 | 4.44 |
ENSMUST00000102920.4
|
Gck
|
glucokinase |
| chr10_+_51367052 | 4.34 |
ENSMUST00000217705.2
ENSMUST00000078778.5 ENSMUST00000220182.2 ENSMUST00000220226.2 |
Lilrb4a
|
leukocyte immunoglobulin-like receptor, subfamily B, member 4A |
| chr1_-_172418058 | 4.33 |
ENSMUST00000065679.8
|
Slamf8
|
SLAM family member 8 |
| chr15_+_77361241 | 4.31 |
ENSMUST00000060551.9
|
Apol10a
|
apolipoprotein L 10A |
| chr19_+_40078132 | 4.28 |
ENSMUST00000068094.13
ENSMUST00000080171.3 |
Cyp2c50
|
cytochrome P450, family 2, subfamily c, polypeptide 50 |
| chr1_-_184578057 | 4.24 |
ENSMUST00000068725.10
|
Mtarc2
|
mitochondrial amidoxime reducing component 2 |
| chr6_+_145067457 | 4.21 |
ENSMUST00000032396.13
|
Lrmp
|
lymphoid-restricted membrane protein |
| chr9_-_45115381 | 4.13 |
ENSMUST00000034599.15
|
Tmprss4
|
transmembrane protease, serine 4 |
| chr8_-_93956143 | 4.13 |
ENSMUST00000176282.2
ENSMUST00000034173.14 |
Ces1e
|
carboxylesterase 1E |
| chr18_+_89215543 | 3.94 |
ENSMUST00000037142.13
|
Cd226
|
CD226 antigen |
| chr1_+_85577709 | 3.93 |
ENSMUST00000155094.8
ENSMUST00000054279.15 ENSMUST00000147552.8 ENSMUST00000153574.8 ENSMUST00000150967.8 |
Sp100
|
nuclear antigen Sp100 |
| chr1_-_156501860 | 3.91 |
ENSMUST00000188964.7
ENSMUST00000190607.2 ENSMUST00000079625.11 |
Tor3a
|
torsin family 3, member A |
| chr4_-_42756489 | 3.89 |
ENSMUST00000140546.3
ENSMUST00000102957.6 |
Ccl19
|
chemokine (C-C motif) ligand 19 |
| chr8_+_93553901 | 3.88 |
ENSMUST00000034187.9
|
Mmp2
|
matrix metallopeptidase 2 |
| chr6_-_125208738 | 3.87 |
ENSMUST00000043422.8
|
Tapbpl
|
TAP binding protein-like |
| chr1_-_85526517 | 3.85 |
ENSMUST00000093508.7
|
Sp110
|
Sp110 nuclear body protein |
| chr8_+_72994152 | 3.83 |
ENSMUST00000126885.2
|
Ap1m1
|
adaptor-related protein complex AP-1, mu subunit 1 |
| chr10_+_116137277 | 3.83 |
ENSMUST00000092167.7
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr13_+_56757389 | 3.81 |
ENSMUST00000045173.10
|
Tgfbi
|
transforming growth factor, beta induced |
| chr7_+_24069680 | 3.75 |
ENSMUST00000205428.2
ENSMUST00000171904.3 ENSMUST00000205626.2 |
Kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr10_+_23727325 | 3.75 |
ENSMUST00000020190.8
|
Vnn3
|
vanin 3 |
| chr18_+_89215438 | 3.71 |
ENSMUST00000237110.2
|
Cd226
|
CD226 antigen |
| chr10_-_75633563 | 3.70 |
ENSMUST00000139724.3
|
Gstt1
|
glutathione S-transferase, theta 1 |
| chr11_-_53750016 | 3.68 |
ENSMUST00000117316.8
ENSMUST00000120776.8 ENSMUST00000121435.2 |
Gm12216
|
predicted gene 12216 |
| chr1_+_85577766 | 3.65 |
ENSMUST00000066427.11
|
Sp100
|
nuclear antigen Sp100 |
| chr13_+_33188511 | 3.65 |
ENSMUST00000006391.5
|
Serpinb9
|
serine (or cysteine) peptidase inhibitor, clade B, member 9 |
| chr7_+_28488380 | 3.64 |
ENSMUST00000209035.2
ENSMUST00000059857.8 |
Rinl
|
Ras and Rab interactor-like |
| chr11_-_82881929 | 3.63 |
ENSMUST00000138797.2
|
Slfn9
|
schlafen 9 |
| chr13_+_24023386 | 3.58 |
ENSMUST00000039721.14
|
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr10_-_86843878 | 3.56 |
ENSMUST00000035288.17
|
Stab2
|
stabilin 2 |
| chr1_-_85248264 | 3.54 |
ENSMUST00000093506.12
ENSMUST00000064341.9 |
C130026I21Rik
|
RIKEN cDNA C130026I21 gene |
| chr3_-_7678796 | 3.52 |
ENSMUST00000192202.6
|
Il7
|
interleukin 7 |
| chr3_-_7678785 | 3.51 |
ENSMUST00000194279.6
|
Il7
|
interleukin 7 |
| chr2_+_43445359 | 3.51 |
ENSMUST00000050511.7
|
Kynu
|
kynureninase |
| chr9_+_38629560 | 3.41 |
ENSMUST00000001544.12
ENSMUST00000118144.8 |
Vwa5a
|
von Willebrand factor A domain containing 5A |
| chr7_-_44785480 | 3.40 |
ENSMUST00000211246.2
ENSMUST00000210197.2 |
Flt3l
|
FMS-like tyrosine kinase 3 ligand |
| chr2_-_153079828 | 3.40 |
ENSMUST00000109795.2
|
Plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr4_-_155012534 | 3.39 |
ENSMUST00000219534.3
|
Tnfrsf14
|
tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) |
| chr5_+_3393893 | 3.38 |
ENSMUST00000165117.8
ENSMUST00000197385.2 |
Cdk6
|
cyclin-dependent kinase 6 |
| chr10_-_116732813 | 3.37 |
ENSMUST00000048229.9
|
Myrfl
|
myelin regulatory factor-like |
| chr4_-_150087587 | 3.25 |
ENSMUST00000084117.13
|
H6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr7_-_101667346 | 3.25 |
ENSMUST00000209844.2
ENSMUST00000211502.2 ENSMUST00000094134.5 |
Il18bp
|
interleukin 18 binding protein |
| chr15_-_76127600 | 3.24 |
ENSMUST00000165738.8
ENSMUST00000075689.7 |
Parp10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr4_-_107928567 | 3.23 |
ENSMUST00000106701.2
|
Scp2
|
sterol carrier protein 2, liver |
| chr11_-_71121559 | 3.22 |
ENSMUST00000108516.9
|
Nlrp1b
|
NLR family, pyrin domain containing 1B |
| chr9_-_106353571 | 3.20 |
ENSMUST00000123555.8
ENSMUST00000125850.2 |
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr18_-_42084249 | 3.17 |
ENSMUST00000070949.6
ENSMUST00000235606.2 |
Prelid2
|
PRELI domain containing 2 |
| chr1_+_153625243 | 3.17 |
ENSMUST00000182722.8
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent) |
| chr14_+_55815879 | 3.16 |
ENSMUST00000174563.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr11_-_70350783 | 3.16 |
ENSMUST00000019064.9
|
Cxcl16
|
chemokine (C-X-C motif) ligand 16 |
| chr1_+_153625161 | 3.15 |
ENSMUST00000086209.10
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent) |
| chr1_+_173248104 | 3.12 |
ENSMUST00000173023.2
|
Aim2
|
absent in melanoma 2 |
| chr12_-_26506422 | 3.12 |
ENSMUST00000020970.10
|
Rsad2
|
radical S-adenosyl methionine domain containing 2 |
| chr8_-_83129134 | 3.11 |
ENSMUST00000209363.2
|
Il15
|
interleukin 15 |
| chr15_-_89263448 | 3.07 |
ENSMUST00000049968.9
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chr15_+_77613348 | 3.06 |
ENSMUST00000230742.2
|
Apol9b
|
apolipoprotein L 9b |
| chr6_-_51989456 | 3.03 |
ENSMUST00000078214.8
ENSMUST00000204778.3 |
Skap2
|
src family associated phosphoprotein 2 |
| chr19_+_12775938 | 3.02 |
ENSMUST00000025601.8
|
Lpxn
|
leupaxin |
| chr2_+_11710523 | 3.01 |
ENSMUST00000138856.2
ENSMUST00000078834.12 ENSMUST00000114834.10 ENSMUST00000114833.10 ENSMUST00000114831.9 |
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr9_-_110745929 | 3.01 |
ENSMUST00000177678.2
|
Fam240a
|
family with sequence similarity 240 member A |
| chr2_+_11710101 | 2.98 |
ENSMUST00000138349.8
ENSMUST00000135341.8 ENSMUST00000128156.9 |
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr8_-_83129832 | 2.97 |
ENSMUST00000034148.7
|
Il15
|
interleukin 15 |
| chr10_+_79660506 | 2.94 |
ENSMUST00000219734.2
|
Misp
|
mitotic spindle positioning |
| chr14_+_55815999 | 2.94 |
ENSMUST00000172738.2
ENSMUST00000089619.13 |
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr16_+_35892437 | 2.88 |
ENSMUST00000163352.9
ENSMUST00000231468.2 |
Ccdc58
|
coiled-coil domain containing 58 |
| chr8_-_85389470 | 2.88 |
ENSMUST00000060427.6
|
Ier2
|
immediate early response 2 |
| chr3_-_95049655 | 2.87 |
ENSMUST00000013851.4
|
Tnfaip8l2
|
tumor necrosis factor, alpha-induced protein 8-like 2 |
| chr7_-_44888220 | 2.86 |
ENSMUST00000210372.2
ENSMUST00000209779.2 ENSMUST00000098461.10 ENSMUST00000211373.2 |
Cd37
|
CD37 antigen |
| chr6_-_70364222 | 2.84 |
ENSMUST00000103392.3
ENSMUST00000195945.2 |
Igkv8-16
|
immunoglobulin kappa variable 8-16 |
| chr11_-_82882613 | 2.84 |
ENSMUST00000092840.11
ENSMUST00000038211.13 |
Slfn9
|
schlafen 9 |
| chr14_+_55815817 | 2.82 |
ENSMUST00000174259.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr18_-_66155651 | 2.81 |
ENSMUST00000143990.2
|
Lman1
|
lectin, mannose-binding, 1 |
| chr1_+_91226058 | 2.80 |
ENSMUST00000027532.13
|
Scly
|
selenocysteine lyase |
| chr11_-_120463667 | 2.78 |
ENSMUST00000168360.2
|
P4hb
|
prolyl 4-hydroxylase, beta polypeptide |
| chr14_-_25927674 | 2.78 |
ENSMUST00000100811.6
|
Tmem254a
|
transmembrane protein 254a |
| chr19_+_55882942 | 2.72 |
ENSMUST00000142291.8
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr14_+_55815580 | 2.71 |
ENSMUST00000174484.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr17_-_30845845 | 2.70 |
ENSMUST00000235547.2
ENSMUST00000188852.2 |
Glo1
1700097N02Rik
|
glyoxalase 1 RIKEN cDNA 1700097N02 gene |
| chr15_+_100202642 | 2.69 |
ENSMUST00000067752.5
ENSMUST00000229588.2 |
Mettl7a1
|
methyltransferase like 7A1 |
| chr15_-_89263790 | 2.66 |
ENSMUST00000238996.2
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chr3_-_86455575 | 2.65 |
ENSMUST00000077524.4
|
Mab21l2
|
mab-21-like 2 |
| chr2_-_105833894 | 2.59 |
ENSMUST00000102555.11
|
Dnajc24
|
DnaJ heat shock protein family (Hsp40) member C24 |
| chr2_-_24809583 | 2.59 |
ENSMUST00000046227.12
ENSMUST00000114432.9 ENSMUST00000091348.11 ENSMUST00000102938.10 ENSMUST00000150379.2 ENSMUST00000152161.8 ENSMUST00000147147.8 |
Ehmt1
|
euchromatic histone methyltransferase 1 |
| chr3_-_101511971 | 2.59 |
ENSMUST00000036493.8
|
Atp1a1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
| chr7_-_4687916 | 2.57 |
ENSMUST00000206306.2
ENSMUST00000205952.2 ENSMUST00000079970.6 |
Hspbp1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr2_+_68966125 | 2.53 |
ENSMUST00000041865.8
|
Nostrin
|
nitric oxide synthase trafficker |
| chr16_-_30086317 | 2.49 |
ENSMUST00000064856.9
|
Cpn2
|
carboxypeptidase N, polypeptide 2 |
| chr1_-_85087567 | 2.48 |
ENSMUST00000161675.3
ENSMUST00000160792.8 |
A530032D15Rik
|
RIKEN cDNA A530032D15Rik gene |
| chr8_-_84059048 | 2.47 |
ENSMUST00000177594.8
ENSMUST00000053902.4 |
Elmod2
|
ELMO/CED-12 domain containing 2 |
| chr2_-_152185901 | 2.41 |
ENSMUST00000040312.7
|
Trib3
|
tribbles pseudokinase 3 |
| chr3_-_9898713 | 2.41 |
ENSMUST00000161949.8
|
Pag1
|
phosphoprotein associated with glycosphingolipid microdomains 1 |
| chr14_+_55909692 | 2.40 |
ENSMUST00000002397.7
|
Gmpr2
|
guanosine monophosphate reductase 2 |
| chr11_-_73215442 | 2.38 |
ENSMUST00000021119.9
|
Aspa
|
aspartoacylase |
| chr9_+_107852733 | 2.36 |
ENSMUST00000035216.11
|
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr1_-_37535170 | 2.35 |
ENSMUST00000148047.2
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr17_+_57369231 | 2.32 |
ENSMUST00000097299.10
ENSMUST00000169543.8 ENSMUST00000163763.2 |
Crb3
|
crumbs family member 3 |
| chr11_-_120464062 | 2.32 |
ENSMUST00000026122.11
|
P4hb
|
prolyl 4-hydroxylase, beta polypeptide |
| chr19_-_3955230 | 2.31 |
ENSMUST00000145791.8
|
Tcirg1
|
T cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 protein A3 |
| chr19_+_34618271 | 2.26 |
ENSMUST00000102824.4
|
Ifit1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chr1_+_171668173 | 2.24 |
ENSMUST00000136479.8
|
Cd84
|
CD84 antigen |
| chr15_-_77527470 | 2.24 |
ENSMUST00000181154.2
ENSMUST00000180949.8 ENSMUST00000166623.10 |
Apol11b
|
apolipoprotein L 11b |
| chr1_+_183766572 | 2.20 |
ENSMUST00000048655.8
|
Dusp10
|
dual specificity phosphatase 10 |
| chr3_-_115800989 | 2.19 |
ENSMUST00000067485.4
|
Slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr2_-_103114105 | 2.19 |
ENSMUST00000111174.8
|
Ehf
|
ets homologous factor |
| chr2_+_11710246 | 2.18 |
ENSMUST00000148748.8
|
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr3_+_63148887 | 2.18 |
ENSMUST00000194324.6
|
Mme
|
membrane metallo endopeptidase |
| chr2_-_51862941 | 2.18 |
ENSMUST00000145481.8
ENSMUST00000112705.9 |
Nmi
|
N-myc (and STAT) interactor |
| chrX_+_7523499 | 2.15 |
ENSMUST00000033485.14
|
Prickle3
|
prickle planar cell polarity protein 3 |
| chr15_-_89263466 | 2.13 |
ENSMUST00000228111.2
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chr6_+_122929627 | 2.13 |
ENSMUST00000204427.2
|
Clec4a3
|
C-type lectin domain family 4, member a3 |
| chr15_+_25843225 | 2.12 |
ENSMUST00000022881.15
|
Retreg1
|
reticulophagy regulator 1 |
| chr16_+_32555015 | 2.11 |
ENSMUST00000239554.1
|
Muc4
|
mucin 4 |
| chr8_-_54091980 | 2.10 |
ENSMUST00000047768.11
|
Neil3
|
nei like 3 (E. coli) |
| chr19_-_5925296 | 2.09 |
ENSMUST00000025728.13
|
Frmd8
|
FERM domain containing 8 |
| chr7_-_101899294 | 2.07 |
ENSMUST00000106923.2
ENSMUST00000098230.11 |
Rhog
|
ras homolog family member G |
| chr5_-_62923463 | 2.07 |
ENSMUST00000076623.8
ENSMUST00000159470.3 |
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr11_+_115778427 | 2.05 |
ENSMUST00000167507.3
|
Myo15b
|
myosin XVB |
| chr2_+_162916551 | 2.02 |
ENSMUST00000142729.3
|
Mybl2
|
myeloblastosis oncogene-like 2 |
| chr5_-_93354348 | 2.01 |
ENSMUST00000058550.15
|
Ccni
|
cyclin I |
| chr11_-_71035530 | 1.95 |
ENSMUST00000108518.3
|
Nlrp1a
|
NLR family, pyrin domain containing 1A |
| chr18_-_46413886 | 1.95 |
ENSMUST00000236999.2
|
Pggt1b
|
protein geranylgeranyltransferase type I, beta subunit |
| chr7_-_44785815 | 1.94 |
ENSMUST00000146760.7
|
Flt3l
|
FMS-like tyrosine kinase 3 ligand |
| chr1_-_37535202 | 1.93 |
ENSMUST00000143636.8
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr3_-_98470822 | 1.92 |
ENSMUST00000186215.7
|
Gm10681
|
predicted gene 10681 |
| chr6_-_130314465 | 1.91 |
ENSMUST00000088017.5
ENSMUST00000111998.9 |
Klra3
|
killer cell lectin-like receptor, subfamily A, member 3 |
| chr3_+_145464413 | 1.91 |
ENSMUST00000029845.15
|
Ddah1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr5_-_93354287 | 1.89 |
ENSMUST00000144514.3
|
Ccni
|
cyclin I |
| chr6_-_99005835 | 1.86 |
ENSMUST00000154163.9
|
Foxp1
|
forkhead box P1 |
| chrX_-_133012457 | 1.86 |
ENSMUST00000159259.3
ENSMUST00000113275.10 |
Nox1
|
NADPH oxidase 1 |
| chr16_-_55858089 | 1.85 |
ENSMUST00000059052.9
|
Trmt10c
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr12_+_8258107 | 1.84 |
ENSMUST00000037383.13
ENSMUST00000218883.2 ENSMUST00000218086.2 ENSMUST00000169104.3 ENSMUST00000217999.2 |
Ldah
|
lipid droplet associated hydrolase |
| chr16_-_97264099 | 1.84 |
ENSMUST00000023655.13
|
Mx1
|
MX dynamin-like GTPase 1 |
| chrX_+_74139645 | 1.84 |
ENSMUST00000131155.8
ENSMUST00000132000.8 |
Dkc1
|
dyskeratosis congenita 1, dyskerin |
| chr13_-_12479804 | 1.83 |
ENSMUST00000124888.8
|
Lgals8
|
lectin, galactose binding, soluble 8 |
| chr7_-_102214636 | 1.82 |
ENSMUST00000106913.3
ENSMUST00000033264.12 |
Trim21
|
tripartite motif-containing 21 |
| chr14_-_55909527 | 1.81 |
ENSMUST00000010520.10
|
Nedd8
|
neural precursor cell expressed, developmentally down-regulated gene 8 |
| chr3_-_9898676 | 1.81 |
ENSMUST00000108384.9
|
Pag1
|
phosphoprotein associated with glycosphingolipid microdomains 1 |
| chr17_+_34138699 | 1.80 |
ENSMUST00000234320.2
|
Tapbp
|
TAP binding protein |
| chr9_-_106353792 | 1.78 |
ENSMUST00000214682.2
ENSMUST00000112479.9 |
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr14_-_73622638 | 1.78 |
ENSMUST00000228637.2
ENSMUST00000022704.9 |
Itm2b
|
integral membrane protein 2B |
| chr14_-_30850881 | 1.77 |
ENSMUST00000203261.3
|
Smim4
|
small integral membrane protein 4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 17.6 | GO:0009087 | methionine catabolic process(GO:0009087) |
| 4.7 | 14.1 | GO:2000547 | regulation of dendritic cell dendrite assembly(GO:2000547) |
| 4.3 | 17.2 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 3.3 | 10.0 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 3.1 | 9.2 | GO:0018900 | dichloromethane metabolic process(GO:0018900) |
| 2.4 | 9.6 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 2.1 | 6.2 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 2.0 | 12.2 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 1.8 | 7.1 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 1.6 | 4.9 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 1.6 | 6.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 1.5 | 10.8 | GO:0090383 | phagosome acidification(GO:0090383) |
| 1.4 | 6.8 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 1.3 | 3.9 | GO:0002590 | negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 1.1 | 3.4 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 1.1 | 3.4 | GO:0036275 | response to 5-fluorouracil(GO:0036275) |
| 1.1 | 6.7 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 1.1 | 7.6 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 1.1 | 13.0 | GO:0045345 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 1.0 | 6.1 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) extrathymic T cell selection(GO:0045062) cellular response to interleukin-15(GO:0071350) |
| 1.0 | 3.0 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 1.0 | 3.0 | GO:0032685 | negative regulation of granulocyte macrophage colony-stimulating factor production(GO:0032685) |
| 1.0 | 5.9 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.9 | 5.6 | GO:0032824 | negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.9 | 4.4 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.9 | 6.1 | GO:0019364 | pyridine nucleotide catabolic process(GO:0019364) |
| 0.9 | 11.2 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.8 | 3.2 | GO:0032382 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 0.8 | 3.2 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 0.8 | 2.4 | GO:0046038 | GMP catabolic process(GO:0046038) |
| 0.8 | 3.1 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.8 | 3.9 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.7 | 9.7 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.7 | 3.7 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.7 | 2.1 | GO:0002838 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) |
| 0.7 | 6.9 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.7 | 5.4 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.7 | 2.0 | GO:0006867 | asparagine transport(GO:0006867) positive regulation of glutamine transport(GO:2000487) |
| 0.6 | 3.2 | GO:0090669 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) telomerase RNA stabilization(GO:0090669) |
| 0.6 | 5.0 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.6 | 1.7 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.5 | 8.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.5 | 2.6 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.5 | 9.8 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.5 | 5.5 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.5 | 8.9 | GO:0032825 | positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.5 | 1.9 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.4 | 1.3 | GO:0045404 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.4 | 0.9 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.4 | 2.2 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.4 | 8.7 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.4 | 12.7 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.4 | 2.8 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.4 | 5.2 | GO:0035646 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.4 | 3.1 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.4 | 4.3 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.4 | 6.0 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.3 | 2.4 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.3 | 7.3 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.3 | 2.4 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.3 | 2.4 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.3 | 2.2 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.3 | 44.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.3 | 6.8 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.3 | 1.8 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.3 | 2.4 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.3 | 4.4 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.3 | 10.1 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.3 | 2.7 | GO:0044334 | canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.3 | 1.9 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.3 | 6.3 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.3 | 1.3 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.3 | 0.8 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.2 | 2.0 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.2 | 1.5 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.2 | 2.1 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.2 | 3.0 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.2 | 3.0 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.2 | 2.3 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.2 | 7.0 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.2 | 3.4 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.2 | 1.5 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.2 | 0.9 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.2 | 1.3 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.2 | 2.3 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.2 | 2.6 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.2 | 1.6 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.2 | 1.0 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.2 | 8.4 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.2 | 0.8 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.2 | 1.8 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.2 | 0.9 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.2 | 2.6 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.2 | 11.8 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.2 | 3.3 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.2 | 2.2 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 1.9 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 3.2 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.1 | 1.4 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.3 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.1 | 0.7 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.1 | 1.4 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 1.7 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.1 | 4.8 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.1 | 0.6 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 2.4 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.1 | 1.3 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 2.6 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.1 | 1.5 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 1.5 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 3.7 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.1 | 1.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 2.7 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 2.2 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 4.8 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.1 | 1.4 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 0.5 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.1 | 0.2 | GO:0015817 | histidine transport(GO:0015817) |
| 0.1 | 2.4 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 2.0 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.1 | 0.4 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.1 | 6.7 | GO:0050868 | negative regulation of T cell activation(GO:0050868) |
| 0.1 | 2.6 | GO:0042088 | T-helper 1 type immune response(GO:0042088) |
| 0.1 | 13.9 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 0.4 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.1 | 0.6 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 | 3.2 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 0.8 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.1 | 3.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 0.2 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.1 | 0.8 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 1.1 | GO:2000696 | regulation of epithelial cell differentiation involved in kidney development(GO:2000696) |
| 0.0 | 2.1 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.0 | 0.5 | GO:0014842 | skeletal muscle satellite cell proliferation(GO:0014841) regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.0 | 1.8 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 1.7 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) negative regulation of telomere capping(GO:1904354) |
| 0.0 | 2.8 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 2.5 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.0 | 1.2 | GO:0032729 | positive regulation of interferon-gamma production(GO:0032729) |
| 0.0 | 1.9 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.5 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.8 | GO:0046519 | sphingoid metabolic process(GO:0046519) |
| 0.0 | 2.9 | GO:0019915 | lipid storage(GO:0019915) |
| 0.0 | 4.3 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.1 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 6.1 | GO:0009308 | amine metabolic process(GO:0009308) |
| 0.0 | 0.5 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.6 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:2000321 | negative regulation of interleukin-1 beta secretion(GO:0050713) positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.0 | 2.0 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 0.0 | 0.9 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 3.0 | GO:0030193 | regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.0 | 3.2 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 2.9 | GO:0071774 | response to fibroblast growth factor(GO:0071774) |
| 0.0 | 1.6 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 | 4.9 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.4 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 0.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 1.4 | GO:0032873 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.0 | 2.6 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 1.1 | GO:0090280 | positive regulation of calcium ion import(GO:0090280) |
| 0.0 | 0.1 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 1.9 | GO:0019395 | fatty acid oxidation(GO:0019395) |
| 0.0 | 0.7 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 1.0 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.1 | GO:0015755 | fructose transport(GO:0015755) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 1.7 | GO:0042098 | T cell proliferation(GO:0042098) |
| 0.0 | 0.4 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.4 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.5 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 1.8 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 1.0 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.0 | 0.7 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.3 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 10.3 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 2.4 | 7.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 1.9 | 11.6 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 1.4 | 8.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 1.1 | 3.4 | GO:0097132 | cyclin D2-CDK6 complex(GO:0097132) |
| 1.0 | 5.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.8 | 3.2 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.8 | 3.2 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.8 | 2.4 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.4 | 4.8 | GO:0045179 | apical cortex(GO:0045179) |
| 0.4 | 8.0 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.4 | 12.9 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.4 | 44.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.4 | 1.1 | GO:0032998 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.3 | 2.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.3 | 2.7 | GO:0042825 | TAP complex(GO:0042825) |
| 0.3 | 1.8 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.3 | 2.7 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.3 | 2.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 4.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 6.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 4.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 10.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.2 | 2.0 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.2 | 2.2 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.2 | 1.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.2 | 2.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 22.0 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 6.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 7.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 14.9 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 1.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 3.2 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 2.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 5.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.7 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 42.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 5.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 5.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.4 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.4 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 8.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 2.1 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.1 | 0.7 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 4.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 5.3 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 0.4 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 1.7 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 2.6 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 1.0 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 22.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 3.0 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 0.6 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 6.8 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 1.8 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 7.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.9 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.2 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 6.2 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 5.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 2.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.5 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 1.1 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 8.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 7.5 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 1.7 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 3.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.9 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 2.0 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 1.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.0 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 1.4 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 2.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.7 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 37.1 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 2.8 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 5.3 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 2.9 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 17.3 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.2 | GO:0032040 | small-subunit processome(GO:0032040) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 17.2 | GO:0004454 | ketohexokinase activity(GO:0004454) |
| 4.4 | 17.6 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 3.1 | 9.2 | GO:0016824 | hydrolase activity, acting on acid halide bonds(GO:0016824) hydrolase activity, acting on acid halide bonds, in C-halide compounds(GO:0019120) alkylhalidase activity(GO:0047651) |
| 2.5 | 10.0 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 1.8 | 5.4 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 1.1 | 3.3 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 1.1 | 4.3 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 1.1 | 11.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 1.0 | 6.2 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 1.0 | 3.9 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.9 | 5.6 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.9 | 2.8 | GO:0070279 | vitamin B6 binding(GO:0070279) |
| 0.9 | 8.0 | GO:0016160 | amylase activity(GO:0016160) |
| 0.9 | 5.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.8 | 3.4 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.8 | 8.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.8 | 3.2 | GO:0033814 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.8 | 2.4 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.8 | 2.4 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.8 | 18.0 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.8 | 6.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.7 | 21.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.7 | 7.0 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.7 | 4.8 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.6 | 1.9 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.6 | 3.0 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) |
| 0.6 | 6.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.5 | 3.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.5 | 2.0 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.5 | 6.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.5 | 3.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.5 | 5.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.5 | 1.8 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) U7 snRNA binding(GO:0071209) |
| 0.4 | 4.4 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.4 | 3.8 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.4 | 4.8 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.4 | 4.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.4 | 2.7 | GO:0046979 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.4 | 1.9 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.4 | 6.7 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.4 | 9.8 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.3 | 45.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.3 | 2.0 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.3 | 3.2 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.3 | 4.5 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.3 | 7.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.3 | 2.4 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.3 | 0.6 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.3 | 2.7 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.3 | 2.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.3 | 4.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 4.7 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.2 | 1.3 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.2 | 2.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) steroid hormone binding(GO:1990239) |
| 0.2 | 7.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.2 | 1.1 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.2 | 1.4 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.2 | 1.9 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 0.7 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.2 | 2.0 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.2 | 2.6 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.2 | 8.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 2.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.4 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 2.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 5.1 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 2.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 3.9 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 2.7 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 2.4 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 1.0 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 0.8 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.1 | 2.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 5.2 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.1 | 0.9 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.2 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.1 | 17.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 8.5 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 1.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 3.4 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.4 | GO:0000010 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.1 | 1.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.5 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.1 | 3.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 4.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 6.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 2.6 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 1.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 2.1 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.1 | 0.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.9 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 2.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 26.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 3.1 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 2.3 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 3.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.4 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 5.8 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.4 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 6.3 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 1.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 5.4 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 3.7 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 1.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 5.4 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 1.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.7 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 3.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 1.8 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 5.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.2 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 2.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.9 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.7 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 7.5 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.9 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.7 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 3.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 13.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 2.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 6.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 3.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 3.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 4.4 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 9.3 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 6.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 5.2 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 7.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 7.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 1.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 10.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 4.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 5.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 11.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 2.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.2 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.2 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 11.0 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.7 | 10.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.5 | 17.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.5 | 1.5 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.4 | 4.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.4 | 7.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.3 | 5.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.3 | 17.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.3 | 19.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.3 | 3.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 21.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.3 | 7.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.3 | 17.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.3 | 7.1 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.2 | 5.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 3.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 4.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 6.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 5.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 2.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 7.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 7.8 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.2 | 8.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.2 | 2.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 4.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 3.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 1.8 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 2.2 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 3.0 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 0.9 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 2.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 3.4 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.1 | 3.3 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 7.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 2.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 3.1 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 2.5 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 1.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 2.1 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 17.1 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.1 | 3.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 2.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 2.8 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 2.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 1.0 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 2.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 2.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.1 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME INTERFERON SIGNALING | Genes involved in Interferon Signaling |