Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
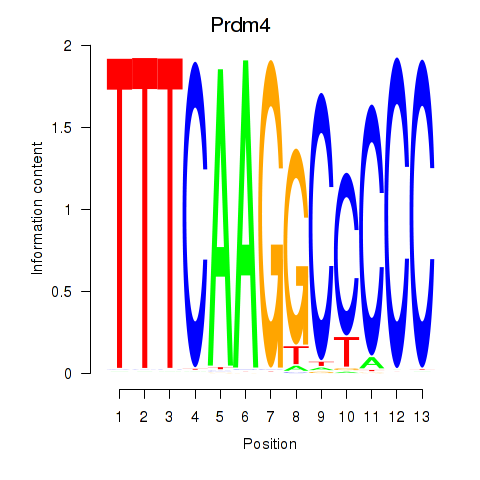
Results for Prdm4
Z-value: 0.56
Transcription factors associated with Prdm4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Prdm4
|
ENSMUSG00000035529.11 | Prdm4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Prdm4 | mm39_v1_chr10_-_85752765_85752870 | 0.08 | 5.2e-01 | Click! |
Activity profile of Prdm4 motif
Sorted Z-values of Prdm4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Prdm4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_113115632 | 7.50 |
ENSMUST00000006523.12
ENSMUST00000200553.2 |
Crip1
|
cysteine-rich protein 1 (intestinal) |
| chrX_+_21581135 | 6.62 |
ENSMUST00000033414.8
|
Slc6a14
|
solute carrier family 6 (neurotransmitter transporter), member 14 |
| chr8_+_123920682 | 5.71 |
ENSMUST00000212409.2
|
Dpep1
|
dipeptidase 1 |
| chr18_+_42186713 | 4.49 |
ENSMUST00000072008.11
|
Sh3rf2
|
SH3 domain containing ring finger 2 |
| chr18_+_42186757 | 4.35 |
ENSMUST00000074679.4
|
Sh3rf2
|
SH3 domain containing ring finger 2 |
| chr11_+_102495189 | 4.32 |
ENSMUST00000057893.7
|
Fzd2
|
frizzled class receptor 2 |
| chr10_+_26698556 | 4.15 |
ENSMUST00000135866.2
|
Arhgap18
|
Rho GTPase activating protein 18 |
| chr10_+_126814542 | 3.77 |
ENSMUST00000105256.10
|
Ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr5_-_142803135 | 3.22 |
ENSMUST00000198181.2
|
Tnrc18
|
trinucleotide repeat containing 18 |
| chr5_-_4808026 | 2.50 |
ENSMUST00000054294.7
|
Fzd1
|
frizzled class receptor 1 |
| chr1_+_74430575 | 2.16 |
ENSMUST00000027367.14
|
Ctdsp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr5_-_142803405 | 2.03 |
ENSMUST00000151477.8
|
Tnrc18
|
trinucleotide repeat containing 18 |
| chr5_-_144902598 | 1.43 |
ENSMUST00000110677.8
ENSMUST00000085684.11 ENSMUST00000100461.7 |
Smurf1
|
SMAD specific E3 ubiquitin protein ligase 1 |
| chr12_-_111947487 | 1.04 |
ENSMUST00000190536.2
|
Rd3l
|
retinal degeneration 3-like |
| chr12_-_111947536 | 0.72 |
ENSMUST00000185354.2
|
Rd3l
|
retinal degeneration 3-like |
| chr12_-_111946560 | 0.38 |
ENSMUST00000190680.2
|
Rd3l
|
retinal degeneration 3-like |
| chr13_-_23735822 | 0.38 |
ENSMUST00000102971.2
|
H4c6
|
H4 clustered histone 6 |
| chr15_-_99372575 | 0.25 |
ENSMUST00000040313.6
|
Bcdin3d
|
BCDIN3 domain containing |
| chr12_+_51424343 | 0.24 |
ENSMUST00000219434.2
ENSMUST00000021335.7 |
Scfd1
|
Sec1 family domain containing 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.7 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 1.0 | 6.8 | GO:0003150 | muscular septum morphogenesis(GO:0003150) |
| 0.5 | 1.4 | GO:0071211 | protein targeting to vacuole involved in autophagy(GO:0071211) |
| 0.5 | 7.5 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.3 | 5.3 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 5.9 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 8.8 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 6.6 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.2 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 4.1 | GO:0030833 | regulation of actin filament polymerization(GO:0030833) |
| 0.0 | 0.2 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.3 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 6.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 5.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.6 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.8 | 5.7 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.6 | 5.9 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.4 | 7.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.3 | 6.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 8.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 5.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.4 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 3.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 1.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 6.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 4.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |