Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Prop1
Z-value: 1.05
Transcription factors associated with Prop1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Prop1
|
ENSMUSG00000044542.4 | Prop1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Prop1 | mm39_v1_chr11_-_50844572_50844596 | -0.16 | 1.8e-01 | Click! |
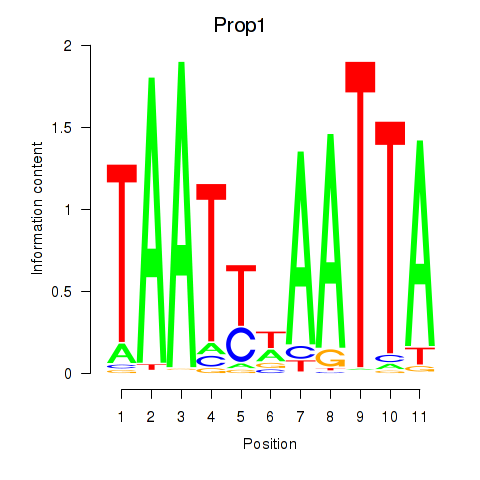
Activity profile of Prop1 motif
Sorted Z-values of Prop1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Prop1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_46597885 | 14.06 |
ENSMUST00000055102.13
ENSMUST00000125008.2 |
Timd2
|
T cell immunoglobulin and mucin domain containing 2 |
| chr7_-_14180496 | 9.27 |
ENSMUST00000063509.11
|
Sult2a8
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
| chr5_-_87682972 | 8.74 |
ENSMUST00000120150.2
|
Sult1b1
|
sulfotransferase family 1B, member 1 |
| chr7_-_14180576 | 8.64 |
ENSMUST00000125941.3
|
Sult2a8
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
| chr3_+_82933383 | 7.90 |
ENSMUST00000029630.15
ENSMUST00000166581.4 |
Fga
|
fibrinogen alpha chain |
| chr19_-_39801188 | 7.77 |
ENSMUST00000162507.2
ENSMUST00000160476.9 ENSMUST00000239028.2 |
Cyp2c40
|
cytochrome P450, family 2, subfamily c, polypeptide 40 |
| chr5_-_89605622 | 7.72 |
ENSMUST00000049209.13
|
Gc
|
vitamin D binding protein |
| chr19_+_39275518 | 7.45 |
ENSMUST00000003137.15
|
Cyp2c29
|
cytochrome P450, family 2, subfamily c, polypeptide 29 |
| chr6_-_115569504 | 7.39 |
ENSMUST00000112957.2
|
Mkrn2os
|
makorin, ring finger protein 2, opposite strand |
| chr1_+_130754413 | 7.17 |
ENSMUST00000027675.14
ENSMUST00000133792.8 |
Pigr
|
polymeric immunoglobulin receptor |
| chr3_+_122688721 | 6.37 |
ENSMUST00000023820.6
|
Fabp2
|
fatty acid binding protein 2, intestinal |
| chr19_-_39637489 | 6.35 |
ENSMUST00000067328.7
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr4_+_98919183 | 6.34 |
ENSMUST00000030280.7
|
Angptl3
|
angiopoietin-like 3 |
| chr19_-_39875192 | 6.09 |
ENSMUST00000168838.3
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69 |
| chr5_-_145521533 | 5.70 |
ENSMUST00000075837.8
|
Cyp3a41b
|
cytochrome P450, family 3, subfamily a, polypeptide 41B |
| chr19_-_40175709 | 5.65 |
ENSMUST00000051846.13
|
Cyp2c70
|
cytochrome P450, family 2, subfamily c, polypeptide 70 |
| chr5_-_145656934 | 5.59 |
ENSMUST00000094111.6
|
Cyp3a41a
|
cytochrome P450, family 3, subfamily a, polypeptide 41A |
| chr6_+_40619913 | 5.56 |
ENSMUST00000238599.2
|
Mgam
|
maltase-glucoamylase |
| chr8_+_13110921 | 5.51 |
ENSMUST00000211363.2
ENSMUST00000033822.4 |
Proz
|
protein Z, vitamin K-dependent plasma glycoprotein |
| chr1_-_130589349 | 5.22 |
ENSMUST00000027657.14
|
C4bp
|
complement component 4 binding protein |
| chrX_-_99638466 | 5.18 |
ENSMUST00000053373.2
|
P2ry4
|
pyrimidinergic receptor P2Y, G-protein coupled, 4 |
| chr1_-_130589321 | 5.07 |
ENSMUST00000137276.3
|
C4bp
|
complement component 4 binding protein |
| chr19_+_38384428 | 4.97 |
ENSMUST00000054098.4
|
Slc35g1
|
solute carrier family 35, member G1 |
| chr8_+_21917427 | 4.91 |
ENSMUST00000095424.6
|
Defa36
|
defensin, alpha, 36 |
| chr13_-_56696310 | 4.90 |
ENSMUST00000062806.6
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr3_+_138121245 | 4.83 |
ENSMUST00000161312.8
ENSMUST00000013458.9 |
Adh4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr3_+_94284739 | 4.81 |
ENSMUST00000197040.5
|
Rorc
|
RAR-related orphan receptor gamma |
| chr8_+_22055402 | 4.73 |
ENSMUST00000084040.3
|
Defa37
|
defensin, alpha, 37 |
| chr19_-_7943365 | 4.69 |
ENSMUST00000182102.8
ENSMUST00000075619.5 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr9_+_78137927 | 4.50 |
ENSMUST00000098537.4
|
Gsta1
|
glutathione S-transferase, alpha 1 (Ya) |
| chr9_+_78164402 | 4.42 |
ENSMUST00000217203.2
|
Gm3776
|
predicted gene 3776 |
| chr3_+_94284812 | 4.33 |
ENSMUST00000200009.2
|
Rorc
|
RAR-related orphan receptor gamma |
| chr14_-_33996185 | 4.33 |
ENSMUST00000227006.2
|
Shld2
|
shieldin complex subunit 2 |
| chr12_-_57592907 | 4.22 |
ENSMUST00000044380.8
|
Foxa1
|
forkhead box A1 |
| chr4_+_115458172 | 4.19 |
ENSMUST00000084342.6
|
Cyp4a32
|
cytochrome P450, family 4, subfamily a, polypeptide 32 |
| chr8_+_21787455 | 4.08 |
ENSMUST00000098892.5
|
Defa5
|
defensin, alpha, 5 |
| chr7_+_100970287 | 3.91 |
ENSMUST00000032927.14
|
Stard10
|
START domain containing 10 |
| chr8_+_22155813 | 3.83 |
ENSMUST00000075268.5
|
Defa34
|
defensin, alpha, 34 |
| chr8_+_21691577 | 3.74 |
ENSMUST00000110754.2
|
Defa41
|
defensin, alpha, 41 |
| chr8_+_21515561 | 3.62 |
ENSMUST00000076754.3
|
Defa21
|
defensin, alpha, 21 |
| chr2_-_110136074 | 3.58 |
ENSMUST00000046233.9
|
Bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase 1 (gamma-butyrobetaine hydroxylase) |
| chr2_-_134396268 | 3.57 |
ENSMUST00000028704.3
|
Hao1
|
hydroxyacid oxidase 1, liver |
| chr17_+_85335775 | 3.33 |
ENSMUST00000024944.9
|
Slc3a1
|
solute carrier family 3, member 1 |
| chr3_+_137923521 | 3.31 |
ENSMUST00000090171.7
|
Adh7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr8_+_21881827 | 3.25 |
ENSMUST00000120874.5
|
Defa33
|
defensin, alpha, 33 |
| chr15_-_96947963 | 3.21 |
ENSMUST00000230907.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr8_+_21868531 | 3.12 |
ENSMUST00000170275.4
|
Defa2
|
defensin, alpha, 2 |
| chr8_+_21999274 | 3.08 |
ENSMUST00000084042.4
|
Defa20
|
defensin, alpha, 20 |
| chr12_+_84098888 | 2.98 |
ENSMUST00000120927.8
ENSMUST00000021653.8 |
Acot3
|
acyl-CoA thioesterase 3 |
| chr4_+_111863441 | 2.97 |
ENSMUST00000162885.8
ENSMUST00000117379.9 ENSMUST00000161389.8 ENSMUST00000162158.2 |
Skint1
|
selection and upkeep of intraepithelial T cells 1 |
| chr4_-_108075119 | 2.94 |
ENSMUST00000223127.2
ENSMUST00000043793.7 ENSMUST00000106690.9 |
Zyg11a
|
zyg-11 family member A, cell cycle regulator |
| chr6_+_41512010 | 2.91 |
ENSMUST00000103288.2
|
Trbj1-5
|
T cell receptor beta joining 1-5 |
| chr8_+_21652293 | 2.87 |
ENSMUST00000098897.2
|
Defa22
|
defensin, alpha, 22 |
| chr6_+_87350292 | 2.82 |
ENSMUST00000032128.6
|
Gkn2
|
gastrokine 2 |
| chr8_+_21555054 | 2.78 |
ENSMUST00000078121.4
|
Defa35
|
defensin, alpha, 35 |
| chrX_-_133442596 | 2.69 |
ENSMUST00000054213.5
|
Timm8a1
|
translocase of inner mitochondrial membrane 8A1 |
| chr13_+_95012107 | 2.63 |
ENSMUST00000022195.13
|
Otp
|
orthopedia homeobox |
| chr14_+_80237691 | 2.59 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr10_+_23727325 | 2.55 |
ENSMUST00000020190.8
|
Vnn3
|
vanin 3 |
| chr8_+_22019048 | 2.52 |
ENSMUST00000084041.4
|
Defa32
|
defensin, alpha, 32 |
| chr3_-_67422821 | 2.51 |
ENSMUST00000054825.5
|
Rarres1
|
retinoic acid receptor responder (tazarotene induced) 1 |
| chr12_-_104439589 | 2.43 |
ENSMUST00000021513.6
|
Gsc
|
goosecoid homeobox |
| chr5_-_87630117 | 2.42 |
ENSMUST00000079811.13
ENSMUST00000144144.3 |
Ugt2a2
|
UDP glucuronosyltransferase 2 family, polypeptide A2 |
| chr4_+_128999325 | 2.41 |
ENSMUST00000106064.10
ENSMUST00000030575.15 ENSMUST00000030577.11 |
Tmem54
|
transmembrane protein 54 |
| chr12_+_9079966 | 2.41 |
ENSMUST00000085741.2
|
Ttc32
|
tetratricopeptide repeat domain 32 |
| chr1_+_134217727 | 2.40 |
ENSMUST00000027730.6
|
Myog
|
myogenin |
| chr12_+_31440842 | 2.39 |
ENSMUST00000167432.8
|
Slc26a3
|
solute carrier family 26, member 3 |
| chr11_-_43792013 | 2.38 |
ENSMUST00000067258.9
ENSMUST00000139906.2 |
Adra1b
|
adrenergic receptor, alpha 1b |
| chr8_-_62576140 | 2.27 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr7_+_101467512 | 2.25 |
ENSMUST00000008090.11
|
Phox2a
|
paired-like homeobox 2a |
| chr17_+_64907697 | 2.22 |
ENSMUST00000086723.10
|
Man2a1
|
mannosidase 2, alpha 1 |
| chr4_-_3938352 | 2.21 |
ENSMUST00000003369.10
|
Plag1
|
pleiomorphic adenoma gene 1 |
| chr11_+_67061908 | 2.08 |
ENSMUST00000018641.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr6_+_86342622 | 2.06 |
ENSMUST00000071492.9
|
Fam136a
|
family with sequence similarity 136, member A |
| chr10_-_115197775 | 2.02 |
ENSMUST00000217848.2
|
Tmem19
|
transmembrane protein 19 |
| chr9_+_66853343 | 1.99 |
ENSMUST00000040917.14
ENSMUST00000127896.8 |
Rps27l
|
ribosomal protein S27-like |
| chr7_+_132212349 | 1.97 |
ENSMUST00000033241.6
ENSMUST00000106170.8 |
Lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr9_+_65538352 | 1.97 |
ENSMUST00000216342.2
ENSMUST00000216382.2 |
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr12_+_9080014 | 1.93 |
ENSMUST00000219488.2
ENSMUST00000219470.2 |
Ttc32
|
tetratricopeptide repeat domain 32 |
| chr11_+_67061837 | 1.89 |
ENSMUST00000170159.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr9_-_53521585 | 1.87 |
ENSMUST00000034547.6
|
Acat1
|
acetyl-Coenzyme A acetyltransferase 1 |
| chr10_-_115198093 | 1.83 |
ENSMUST00000219890.2
ENSMUST00000218731.2 ENSMUST00000217887.2 ENSMUST00000092170.7 |
Tmem19
|
transmembrane protein 19 |
| chr6_+_41511733 | 1.78 |
ENSMUST00000103287.2
|
Trbj1-4
|
T cell receptor beta joining 1-4 |
| chr13_+_24511387 | 1.76 |
ENSMUST00000224953.2
ENSMUST00000050859.13 ENSMUST00000167746.8 ENSMUST00000224819.2 |
Cmah
|
cytidine monophospho-N-acetylneuraminic acid hydroxylase |
| chr11_-_117764258 | 1.74 |
ENSMUST00000033230.8
|
Tha1
|
threonine aldolase 1 |
| chr4_+_95445731 | 1.74 |
ENSMUST00000079223.11
ENSMUST00000177394.8 |
Fggy
|
FGGY carbohydrate kinase domain containing |
| chr14_-_48904998 | 1.71 |
ENSMUST00000152018.8
|
Otx2
|
orthodenticle homeobox 2 |
| chr7_-_86016045 | 1.67 |
ENSMUST00000213255.2
ENSMUST00000216700.2 ENSMUST00000213869.2 |
Olfr305
|
olfactory receptor 305 |
| chr13_-_19521337 | 1.66 |
ENSMUST00000103563.3
|
Trgv2
|
T cell receptor gamma variable 2 |
| chr7_+_126895531 | 1.66 |
ENSMUST00000170971.8
|
Itgal
|
integrin alpha L |
| chr14_+_73475335 | 1.61 |
ENSMUST00000044405.8
|
Lpar6
|
lysophosphatidic acid receptor 6 |
| chr12_+_59176543 | 1.61 |
ENSMUST00000069430.15
ENSMUST00000177370.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr10_+_79977291 | 1.60 |
ENSMUST00000105367.8
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr1_+_106866678 | 1.59 |
ENSMUST00000112724.3
|
Serpinb12
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 12 |
| chr11_-_59054521 | 1.58 |
ENSMUST00000137433.2
ENSMUST00000054523.6 |
Iba57
|
IBA57 homolog, iron-sulfur cluster assembly |
| chr7_+_126895463 | 1.51 |
ENSMUST00000106306.9
ENSMUST00000120857.8 |
Itgal
|
integrin alpha L |
| chr17_+_38104420 | 1.51 |
ENSMUST00000216051.3
|
Olfr123
|
olfactory receptor 123 |
| chr12_-_113958518 | 1.50 |
ENSMUST00000103467.2
|
Ighv14-2
|
immunoglobulin heavy variable 14-2 |
| chr7_+_126895423 | 1.50 |
ENSMUST00000117762.8
|
Itgal
|
integrin alpha L |
| chr14_-_48904958 | 1.49 |
ENSMUST00000144465.8
ENSMUST00000133479.8 ENSMUST00000119070.8 ENSMUST00000226501.2 |
Otx2
|
orthodenticle homeobox 2 |
| chr3_+_84500854 | 1.47 |
ENSMUST00000062623.4
|
Tigd4
|
tigger transposable element derived 4 |
| chr6_+_41511248 | 1.47 |
ENSMUST00000192366.2
ENSMUST00000103286.2 |
Trbj1-3
|
T cell receptor beta joining 1-3 |
| chr16_-_89368059 | 1.46 |
ENSMUST00000171542.2
|
Krtap11-1
|
keratin associated protein 11-1 |
| chr12_+_59176506 | 1.45 |
ENSMUST00000175912.8
ENSMUST00000176892.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr1_+_167426019 | 1.44 |
ENSMUST00000111386.8
ENSMUST00000111384.8 |
Rxrg
|
retinoid X receptor gamma |
| chr17_-_45910529 | 1.41 |
ENSMUST00000171847.8
ENSMUST00000166633.8 ENSMUST00000169729.8 |
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr4_-_82423985 | 1.40 |
ENSMUST00000107245.9
ENSMUST00000107246.2 |
Nfib
|
nuclear factor I/B |
| chr19_-_45224251 | 1.39 |
ENSMUST00000099401.6
|
Lbx1
|
ladybird homeobox 1 |
| chr18_-_9282754 | 1.37 |
ENSMUST00000041007.4
|
Gjd4
|
gap junction protein, delta 4 |
| chr15_-_101651534 | 1.36 |
ENSMUST00000023710.6
|
Krt71
|
keratin 71 |
| chr1_+_6805048 | 1.35 |
ENSMUST00000139838.8
|
St18
|
suppression of tumorigenicity 18 |
| chr18_+_31742565 | 1.34 |
ENSMUST00000164667.2
|
B930094E09Rik
|
RIKEN cDNA B930094E09 gene |
| chr11_+_100978103 | 1.33 |
ENSMUST00000107302.8
ENSMUST00000107303.10 ENSMUST00000017945.15 ENSMUST00000149597.2 |
Mlx
|
MAX-like protein X |
| chr2_+_88505972 | 1.31 |
ENSMUST00000216767.2
ENSMUST00000213893.2 |
Olfr1193
|
olfactory receptor 1193 |
| chr1_+_167425953 | 1.30 |
ENSMUST00000015987.10
|
Rxrg
|
retinoid X receptor gamma |
| chr7_+_140521450 | 1.26 |
ENSMUST00000164580.3
ENSMUST00000079403.11 |
Pgghg
|
protein glucosylgalactosylhydroxylysine glucosidase |
| chr12_-_113928438 | 1.26 |
ENSMUST00000103478.4
|
Ighv3-1
|
immunoglobulin heavy variable 3-1 |
| chr13_-_3968157 | 1.26 |
ENSMUST00000223258.2
ENSMUST00000091853.12 |
Net1
|
neuroepithelial cell transforming gene 1 |
| chr6_+_42117870 | 1.26 |
ENSMUST00000057686.5
|
Tas2r139
|
taste receptor, type 2, member 139 |
| chr4_+_47474715 | 1.26 |
ENSMUST00000137461.8
ENSMUST00000125622.2 |
Sec61b
|
Sec61 beta subunit |
| chr14_+_52964069 | 1.22 |
ENSMUST00000184883.2
|
Trav6d-3
|
T cell receptor alpha variable 6D-3 |
| chr1_+_172777976 | 1.22 |
ENSMUST00000215254.2
|
Olfr16
|
olfactory receptor 16 |
| chr3_-_84063067 | 1.21 |
ENSMUST00000047368.8
|
Mnd1
|
meiotic nuclear divisions 1 |
| chr5_+_114268425 | 1.20 |
ENSMUST00000031587.13
|
Ung
|
uracil DNA glycosylase |
| chr6_+_139564196 | 1.20 |
ENSMUST00000188066.2
ENSMUST00000190962.7 |
Pik3c2g
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
| chr18_-_14105591 | 1.20 |
ENSMUST00000234025.2
|
Zfp521
|
zinc finger protein 521 |
| chr9_+_53212871 | 1.20 |
ENSMUST00000051014.2
|
Exph5
|
exophilin 5 |
| chr14_+_52904815 | 1.20 |
ENSMUST00000198058.2
ENSMUST00000103573.3 |
Trav6-2
|
T cell receptor alpha variable 6-2 |
| chrX_-_93256244 | 1.19 |
ENSMUST00000113922.2
|
Eif2s3x
|
eukaryotic translation initiation factor 2, subunit 3, structural gene X-linked |
| chr2_-_72817060 | 1.19 |
ENSMUST00000112062.2
|
Gm11084
|
predicted gene 11084 |
| chr15_-_63680596 | 1.16 |
ENSMUST00000110125.9
ENSMUST00000173503.3 |
Gsdmc
|
gasdermin C |
| chr4_+_47474652 | 1.15 |
ENSMUST00000065678.6
|
Sec61b
|
Sec61 beta subunit |
| chr12_+_85157607 | 1.15 |
ENSMUST00000053811.10
|
Dlst
|
dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) |
| chr2_-_89671899 | 1.14 |
ENSMUST00000213833.2
|
Olfr1256
|
olfactory receptor 1256 |
| chr2_-_71377088 | 1.11 |
ENSMUST00000024159.8
|
Dlx2
|
distal-less homeobox 2 |
| chr15_-_56557920 | 1.11 |
ENSMUST00000050544.8
|
Has2
|
hyaluronan synthase 2 |
| chr19_-_13807805 | 1.09 |
ENSMUST00000214475.2
ENSMUST00000067670.5 ENSMUST00000219674.2 ENSMUST00000216287.2 ENSMUST00000215760.2 |
Olfr1500
|
olfactory receptor 1500 |
| chr8_-_65582206 | 1.07 |
ENSMUST00000098713.5
|
Smim31
|
small integral membrane protein 31 |
| chr4_+_109835224 | 1.07 |
ENSMUST00000061187.4
|
Dmrta2
|
doublesex and mab-3 related transcription factor like family A2 |
| chr10_-_128996851 | 1.06 |
ENSMUST00000078914.3
|
Olfr771
|
olfactory receptor 771 |
| chr5_+_42225303 | 1.05 |
ENSMUST00000087332.5
|
Gm16223
|
predicted gene 16223 |
| chr12_+_79344056 | 1.04 |
ENSMUST00000171210.3
|
Rad51b
|
RAD51 paralog B |
| chr16_+_65317389 | 1.04 |
ENSMUST00000176330.8
ENSMUST00000004964.15 ENSMUST00000176038.8 |
Pou1f1
|
POU domain, class 1, transcription factor 1 |
| chr19_-_13828056 | 1.02 |
ENSMUST00000208493.3
|
Olfr1501
|
olfactory receptor 1501 |
| chr9_-_48876290 | 1.02 |
ENSMUST00000008734.5
|
Htr3b
|
5-hydroxytryptamine (serotonin) receptor 3B |
| chr1_+_37911415 | 1.01 |
ENSMUST00000142670.2
|
Lipt1
|
lipoyltransferase 1 |
| chr6_+_3993774 | 1.00 |
ENSMUST00000031673.7
|
Gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr13_+_19524136 | 1.00 |
ENSMUST00000103564.3
|
Trgv1
|
T cell receptor gamma, variable 1 |
| chr6_-_5298455 | 0.99 |
ENSMUST00000057792.9
|
Pon2
|
paraoxonase 2 |
| chr11_+_84020475 | 0.98 |
ENSMUST00000133811.3
|
Acaca
|
acetyl-Coenzyme A carboxylase alpha |
| chr1_+_187730018 | 0.97 |
ENSMUST00000027906.13
|
Esrrg
|
estrogen-related receptor gamma |
| chrX_+_109857866 | 0.97 |
ENSMUST00000078229.5
|
Pou3f4
|
POU domain, class 3, transcription factor 4 |
| chr6_-_113411718 | 0.96 |
ENSMUST00000113091.8
|
Cidec
|
cell death-inducing DFFA-like effector c |
| chrX_+_36915787 | 0.96 |
ENSMUST00000006362.4
|
Rhox6
|
reproductive homeobox 6 |
| chr1_-_163552693 | 0.96 |
ENSMUST00000159679.8
|
Mettl11b
|
methyltransferase like 11B |
| chr17_+_27775613 | 0.96 |
ENSMUST00000231780.2
ENSMUST00000232253.2 ENSMUST00000232552.2 ENSMUST00000117600.9 |
Hmga1
|
high mobility group AT-hook 1 |
| chrX_-_36990581 | 0.95 |
ENSMUST00000170643.4
ENSMUST00000089062.8 |
Rhox9
|
reproductive homeobox 9 |
| chr6_-_83098255 | 0.94 |
ENSMUST00000205023.2
ENSMUST00000146328.4 ENSMUST00000151393.7 ENSMUST00000032111.11 ENSMUST00000113936.10 |
Wbp1
|
WW domain binding protein 1 |
| chrX_+_55939732 | 0.94 |
ENSMUST00000136396.8
|
Adgrg4
|
adhesion G protein-coupled receptor G4 |
| chr15_-_103929556 | 0.94 |
ENSMUST00000226584.2
ENSMUST00000037685.9 |
Mucl2
|
mucin-like 2 |
| chr19_+_12651543 | 0.93 |
ENSMUST00000057924.3
|
Olfr1442
|
olfactory receptor 1442 |
| chrX_+_37689503 | 0.93 |
ENSMUST00000000365.3
|
Mcts1
|
malignant T cell amplified sequence 1 |
| chr17_+_35878332 | 0.92 |
ENSMUST00000044326.5
|
2300002M23Rik
|
RIKEN cDNA 2300002M23 gene |
| chr11_+_43046476 | 0.91 |
ENSMUST00000238415.2
|
Atp10b
|
ATPase, class V, type 10B |
| chr15_+_53748302 | 0.91 |
ENSMUST00000100666.3
|
Gm7489
|
predicted gene 7489 |
| chr14_+_64229914 | 0.91 |
ENSMUST00000058229.6
|
Rp1l1
|
retinitis pigmentosa 1 homolog like 1 |
| chrX_+_37861548 | 0.91 |
ENSMUST00000050744.6
|
6030498E09Rik
|
RIKEN cDNA 6030498E09 gene |
| chr14_+_53941464 | 0.91 |
ENSMUST00000103664.6
|
Trav5-4
|
T cell receptor alpha variable 5-4 |
| chr2_-_86317730 | 0.90 |
ENSMUST00000217292.3
|
Olfr228
|
olfactory receptor 228 |
| chr17_+_37769807 | 0.90 |
ENSMUST00000214668.2
ENSMUST00000217602.2 ENSMUST00000214938.2 |
Olfr109
|
olfactory receptor 109 |
| chr14_+_73411249 | 0.90 |
ENSMUST00000166875.2
|
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr3_-_116506294 | 0.89 |
ENSMUST00000029569.9
|
Slc35a3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member 3 |
| chr14_+_54436247 | 0.89 |
ENSMUST00000103720.2
|
Traj21
|
T cell receptor alpha joining 21 |
| chr14_-_51433380 | 0.88 |
ENSMUST00000051274.2
|
Ang2
|
angiogenin, ribonuclease A family, member 2 |
| chrX_-_167970196 | 0.87 |
ENSMUST00000066112.12
ENSMUST00000112118.8 ENSMUST00000112120.8 ENSMUST00000112119.8 |
Amelx
|
amelogenin, X-linked |
| chr8_+_124204598 | 0.86 |
ENSMUST00000001520.13
|
Afg3l1
|
AFG3-like AAA ATPase 1 |
| chr1_+_187730032 | 0.86 |
ENSMUST00000110938.2
|
Esrrg
|
estrogen-related receptor gamma |
| chrM_+_10167 | 0.86 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr6_+_63232955 | 0.86 |
ENSMUST00000095852.5
|
Grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr3_-_72875187 | 0.86 |
ENSMUST00000167334.8
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr3_+_122213420 | 0.85 |
ENSMUST00000029766.9
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr2_-_88848668 | 0.84 |
ENSMUST00000216000.2
ENSMUST00000217000.2 |
Olfr1216
|
olfactory receptor 1216 |
| chr1_+_58750647 | 0.83 |
ENSMUST00000097722.9
ENSMUST00000114313.8 |
Cflar
|
CASP8 and FADD-like apoptosis regulator |
| chr7_+_19738020 | 0.82 |
ENSMUST00000137183.9
|
Nlrp9b
|
NLR family, pyrin domain containing 9B |
| chr2_-_155787197 | 0.81 |
ENSMUST00000040162.3
|
Gdf5
|
growth differentiation factor 5 |
| chr17_+_27775637 | 0.81 |
ENSMUST00000117254.9
ENSMUST00000231243.2 ENSMUST00000231358.2 ENSMUST00000118570.2 ENSMUST00000231796.2 |
Hmga1
|
high mobility group AT-hook 1 |
| chr6_-_70237939 | 0.81 |
ENSMUST00000103386.3
|
Igkv6-23
|
immunoglobulin kappa variable 6-23 |
| chr6_+_149043011 | 0.81 |
ENSMUST00000179873.8
ENSMUST00000047531.16 ENSMUST00000111548.8 ENSMUST00000111547.2 ENSMUST00000134306.8 ENSMUST00000147934.4 |
Etfbkmt
|
electron transfer flavoprotein beta subunit lysine methyltransferase |
| chr7_-_102540089 | 0.80 |
ENSMUST00000217024.2
|
Olfr569
|
olfactory receptor 569 |
| chr11_+_101932328 | 0.80 |
ENSMUST00000123895.8
ENSMUST00000017453.12 ENSMUST00000107163.9 ENSMUST00000107164.3 |
Cd300lg
|
CD300 molecule like family member G |
| chr7_+_106740521 | 0.79 |
ENSMUST00000210474.2
|
Olfr716
|
olfactory receptor 716 |
| chr3_+_103646682 | 0.79 |
ENSMUST00000106852.2
|
Gm10964
|
predicted gene 10964 |
| chr2_+_150751475 | 0.79 |
ENSMUST00000028948.5
|
Gins1
|
GINS complex subunit 1 (Psf1 homolog) |
| chr5_+_104318542 | 0.79 |
ENSMUST00000112771.2
|
Dspp
|
dentin sialophosphoprotein |
| chr5_+_92285748 | 0.79 |
ENSMUST00000031355.10
ENSMUST00000202155.2 |
Uso1
|
USO1 vesicle docking factor |
| chr6_-_70412460 | 0.79 |
ENSMUST00000103394.2
|
Igkv6-14
|
immunoglobulin kappa variable 6-14 |
| chrX_-_133177717 | 0.77 |
ENSMUST00000087541.12
ENSMUST00000087540.4 |
Trmt2b
|
TRM2 tRNA methyltransferase 2B |
| chr1_+_174196485 | 0.77 |
ENSMUST00000085862.2
|
Olfr417
|
olfactory receptor 417 |
| chr18_-_70409277 | 0.77 |
ENSMUST00000239144.2
|
Gm36255
|
predicted gene, 36255 |
| chr9_+_39777718 | 0.75 |
ENSMUST00000215303.3
ENSMUST00000216167.2 |
Olfr972
|
olfactory receptor 972 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.0 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 2.4 | 16.9 | GO:0006067 | ethanol metabolic process(GO:0006067) |
| 2.0 | 7.9 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 1.4 | 4.2 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 1.2 | 3.6 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 1.1 | 6.3 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 1.0 | 32.9 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 1.0 | 3.0 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.9 | 3.6 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.8 | 9.1 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.8 | 2.4 | GO:0014737 | positive regulation of muscle atrophy(GO:0014737) response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.8 | 2.4 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.8 | 2.3 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.7 | 5.0 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.6 | 1.9 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.6 | 2.4 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.6 | 5.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.5 | 4.7 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.5 | 1.5 | GO:1904736 | negative regulation of electron carrier activity(GO:1904733) regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904735) negative regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904736) |
| 0.5 | 1.9 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.5 | 3.2 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.5 | 1.4 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.4 | 1.8 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.4 | 1.8 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.4 | 0.4 | GO:1904444 | regulation of establishment of Sertoli cell barrier(GO:1904444) |
| 0.4 | 1.2 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.4 | 3.9 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.4 | 7.7 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.4 | 1.1 | GO:0021893 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.4 | 1.1 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.4 | 14.1 | GO:0097286 | iron ion import(GO:0097286) |
| 0.3 | 1.7 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.3 | 4.7 | GO:0015747 | urate transport(GO:0015747) |
| 0.3 | 1.5 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.2 | 2.4 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.2 | 4.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.2 | 1.4 | GO:0014901 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.2 | 2.4 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.2 | 1.9 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.2 | 0.8 | GO:0014732 | skeletal muscle atrophy(GO:0014732) negative regulation of myoblast fusion(GO:1901740) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.2 | 1.4 | GO:0015862 | uridine transport(GO:0015862) |
| 0.2 | 2.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 1.0 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.2 | 2.6 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.2 | 0.7 | GO:1903999 | negative regulation of eating behavior(GO:1903999) regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.2 | 1.2 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.2 | 6.4 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.2 | 0.5 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.2 | 2.9 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 | 4.5 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.1 | 0.6 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 1.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 2.0 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 1.4 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 0.6 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 0.8 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.1 | 0.9 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 1.7 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.1 | 0.3 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.4 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.1 | 0.6 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 0.5 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.8 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.1 | 0.4 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.1 | 0.3 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.1 | 3.1 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 0.7 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.1 | 0.5 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 5.5 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 13.1 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 1.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 1.8 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 2.2 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.1 | 5.2 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 1.3 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 | 0.2 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.1 | 3.1 | GO:0070328 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.1 | 2.6 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.1 | 0.2 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 0.4 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 2.0 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 1.3 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 3.2 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 1.0 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.1 | 0.7 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) |
| 0.1 | 0.8 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 13.9 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.1 | 0.9 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 2.4 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.1 | GO:1990428 | miRNA transport(GO:1990428) |
| 0.0 | 0.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 1.6 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 1.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.5 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 2.9 | GO:0051438 | regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.0 | 0.5 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 1.0 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 2.6 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 1.1 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 | 1.0 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.3 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.4 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 1.2 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 1.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.9 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 0.1 | GO:0009726 | detection of nodal flow(GO:0003127) detection of endogenous stimulus(GO:0009726) |
| 0.0 | 1.2 | GO:0035825 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 1.0 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 0.1 | GO:0060082 | response to carbon monoxide(GO:0034465) eye blink reflex(GO:0060082) |
| 0.0 | 0.8 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.7 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.3 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.0 | 0.4 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.8 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.7 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.0 | 0.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 4.8 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 14.8 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.9 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.1 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.4 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.1 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 3.0 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.0 | GO:0046333 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.0 | 0.4 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 2.2 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.2 | GO:2000252 | negative regulation of feeding behavior(GO:2000252) |
| 0.0 | 0.8 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.1 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 1.5 | GO:0008033 | tRNA processing(GO:0008033) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.7 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.9 | 7.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.6 | 6.4 | GO:0045179 | apical cortex(GO:0045179) |
| 0.5 | 4.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.3 | 2.4 | GO:0031205 | Sec61 translocon complex(GO:0005784) endoplasmic reticulum Sec complex(GO:0031205) translocon complex(GO:0071256) |
| 0.3 | 1.0 | GO:1904602 | serotonin-activated cation-selective channel complex(GO:1904602) |
| 0.3 | 2.4 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.3 | 0.8 | GO:0000811 | GINS complex(GO:0000811) |
| 0.3 | 1.3 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.2 | 0.9 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.2 | 0.6 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.2 | 3.0 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.2 | 1.6 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.2 | 0.7 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.2 | 1.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.2 | 0.5 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.2 | 7.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 1.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 2.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 1.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 2.6 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 0.4 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 0.5 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 1.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.8 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 1.9 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 3.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.6 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 5.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.8 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 10.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 2.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 2.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 4.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 5.0 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 12.0 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.6 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 1.7 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 5.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 4.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 5.9 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.0 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.0 | 4.1 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 14.1 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 2.7 | 8.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 1.9 | 7.7 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 1.9 | 9.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 1.7 | 5.2 | GO:0045030 | UTP-activated nucleotide receptor activity(GO:0045030) |
| 1.5 | 9.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 1.4 | 11.3 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 1.2 | 7.5 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.9 | 4.7 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.9 | 29.7 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.9 | 5.6 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.9 | 3.6 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.9 | 0.9 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.7 | 2.2 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.7 | 6.4 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.6 | 7.2 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.6 | 2.4 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.6 | 1.8 | GO:0030338 | CMP-N-acetylneuraminate monooxygenase activity(GO:0030338) |
| 0.6 | 1.7 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.6 | 6.3 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.5 | 2.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.4 | 1.3 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.4 | 1.7 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.4 | 2.4 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.3 | 1.2 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.3 | 0.9 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.3 | 1.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.3 | 1.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.3 | 4.7 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.3 | 2.7 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.3 | 1.9 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.2 | 1.5 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.2 | 2.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.2 | 1.6 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 0.6 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.2 | 1.0 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.2 | 1.0 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.2 | 1.0 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.2 | 2.0 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.2 | 1.8 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.2 | 4.5 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.1 | 3.0 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 2.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 1.3 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.1 | 0.5 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.6 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.5 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 1.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.2 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 0.1 | 0.8 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.7 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 0.4 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 1.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.8 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 1.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.8 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 2.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.5 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.1 | 0.2 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 1.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 1.4 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.7 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.8 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.4 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.7 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 2.0 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 2.5 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 2.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.3 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.5 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 5.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 3.0 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 5.1 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.6 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 1.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 2.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 21.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.3 | GO:0004549 | tRNA-specific ribonuclease activity(GO:0004549) |
| 0.0 | 0.1 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.0 | 0.0 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.1 | GO:1990247 | pre-mRNA intronic binding(GO:0097157) N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.0 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 12.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 6.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 4.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 0.8 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 2.7 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 6.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 6.5 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 2.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.3 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 2.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.7 | 8.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.6 | 8.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.6 | 5.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.5 | 7.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.3 | 2.4 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.3 | 4.1 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.3 | 4.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 7.7 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.0 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 13.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.3 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.1 | 6.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 4.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 2.2 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 1.9 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 1.1 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 2.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.8 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 7.2 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 2.4 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 1.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.2 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.0 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 1.0 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 2.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 1.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |