Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Prox1
Z-value: 0.81
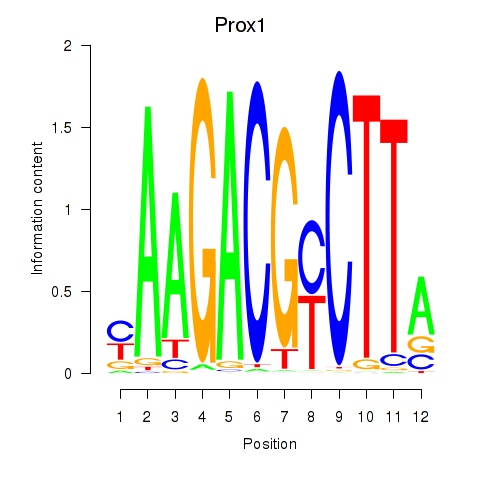
Transcription factors associated with Prox1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Prox1
|
ENSMUSG00000010175.14 | Prox1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Prox1 | mm39_v1_chr1_-_189901596_189901723 | -0.16 | 1.7e-01 | Click! |
Activity profile of Prox1 motif
Sorted Z-values of Prox1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Prox1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_22965286 | 5.69 |
ENSMUST00000023593.6
|
Adipoq
|
adiponectin, C1Q and collagen domain containing |
| chr16_+_22965330 | 5.04 |
ENSMUST00000171309.2
|
Adipoq
|
adiponectin, C1Q and collagen domain containing |
| chr19_-_15901919 | 4.38 |
ENSMUST00000162053.8
|
Psat1
|
phosphoserine aminotransferase 1 |
| chr10_-_80649315 | 4.19 |
ENSMUST00000181039.8
ENSMUST00000180438.2 |
Jsrp1
|
junctional sarcoplasmic reticulum protein 1 |
| chr7_+_141995545 | 4.07 |
ENSMUST00000105971.8
ENSMUST00000145287.8 |
Tnni2
|
troponin I, skeletal, fast 2 |
| chr18_-_68562385 | 3.90 |
ENSMUST00000052347.8
|
Mc2r
|
melanocortin 2 receptor |
| chr14_-_56181993 | 3.89 |
ENSMUST00000022834.7
ENSMUST00000226280.2 |
Cma1
|
chymase 1, mast cell |
| chr4_-_137157824 | 3.83 |
ENSMUST00000102522.5
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr17_+_29538157 | 3.70 |
ENSMUST00000114699.9
ENSMUST00000155348.3 ENSMUST00000234618.2 |
Pi16
|
peptidase inhibitor 16 |
| chr14_-_51384236 | 3.56 |
ENSMUST00000080126.4
|
Rnase1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr15_+_102898966 | 3.47 |
ENSMUST00000001703.8
|
Hoxc8
|
homeobox C8 |
| chr11_+_4936824 | 2.59 |
ENSMUST00000109897.8
ENSMUST00000009234.16 |
Ap1b1
|
adaptor protein complex AP-1, beta 1 subunit |
| chr4_+_42950367 | 2.42 |
ENSMUST00000084662.12
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr1_-_169575203 | 2.24 |
ENSMUST00000027991.12
ENSMUST00000111357.2 |
Rgs4
|
regulator of G-protein signaling 4 |
| chr8_+_31601837 | 2.11 |
ENSMUST00000046941.8
ENSMUST00000217278.2 |
Rnf122
|
ring finger protein 122 |
| chr5_+_24305577 | 1.91 |
ENSMUST00000030841.10
ENSMUST00000163409.5 |
Klhl7
|
kelch-like 7 |
| chr15_-_86070338 | 1.90 |
ENSMUST00000044332.16
|
Cerk
|
ceramide kinase |
| chrY_-_90850446 | 1.83 |
ENSMUST00000179623.2
|
Gm21748
|
predicted gene, 21748 |
| chr4_+_130640436 | 1.80 |
ENSMUST00000151698.8
|
Laptm5
|
lysosomal-associated protein transmembrane 5 |
| chr8_-_70929555 | 1.79 |
ENSMUST00000066597.13
ENSMUST00000210250.2 ENSMUST00000209415.2 ENSMUST00000166976.3 |
Klhl26
|
kelch-like 26 |
| chr4_+_130640611 | 1.77 |
ENSMUST00000156225.8
ENSMUST00000156742.8 |
Laptm5
|
lysosomal-associated protein transmembrane 5 |
| chr17_+_37356854 | 1.76 |
ENSMUST00000025338.16
|
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr18_-_67378886 | 1.61 |
ENSMUST00000073054.5
|
Mppe1
|
metallophosphoesterase 1 |
| chr6_+_146697539 | 1.58 |
ENSMUST00000111639.8
|
Arntl2
|
aryl hydrocarbon receptor nuclear translocator-like 2 |
| chr8_+_89015705 | 1.44 |
ENSMUST00000171456.9
|
Adcy7
|
adenylate cyclase 7 |
| chr6_-_131365380 | 1.44 |
ENSMUST00000032309.13
ENSMUST00000087865.4 |
Ybx3
|
Y box protein 3 |
| chr4_-_133694607 | 1.40 |
ENSMUST00000105893.8
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr18_+_37864045 | 1.32 |
ENSMUST00000192535.2
|
Pcdhgb5
|
protocadherin gamma subfamily B, 5 |
| chr5_+_139238069 | 1.31 |
ENSMUST00000026976.12
|
Get4
|
golgi to ER traffic protein 4 |
| chr5_+_139197689 | 1.30 |
ENSMUST00000148772.8
ENSMUST00000110882.8 |
Sun1
|
Sad1 and UNC84 domain containing 1 |
| chr17_+_37356872 | 1.27 |
ENSMUST00000174456.8
|
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr8_+_61085890 | 1.27 |
ENSMUST00000160719.8
|
Mfap3l
|
microfibrillar-associated protein 3-like |
| chr4_+_74160705 | 1.25 |
ENSMUST00000077851.10
|
Kdm4c
|
lysine (K)-specific demethylase 4C |
| chr1_+_78794475 | 1.20 |
ENSMUST00000057262.8
ENSMUST00000187432.2 |
Kcne4
|
potassium voltage-gated channel, Isk-related subfamily, gene 4 |
| chr18_-_67378512 | 1.17 |
ENSMUST00000237631.2
|
Mppe1
|
metallophosphoesterase 1 |
| chr4_-_137137088 | 1.14 |
ENSMUST00000024200.7
|
Cela3a
|
chymotrypsin-like elastase family, member 3A |
| chr17_+_28059129 | 1.13 |
ENSMUST00000233657.2
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr4_-_133694543 | 1.10 |
ENSMUST00000123234.8
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr5_-_124492734 | 1.08 |
ENSMUST00000031341.11
|
Cdk2ap1
|
CDK2 (cyclin-dependent kinase 2)-associated protein 1 |
| chr17_-_35827676 | 1.06 |
ENSMUST00000160885.2
ENSMUST00000159009.2 ENSMUST00000161012.8 |
Tcf19
|
transcription factor 19 |
| chr17_-_33613544 | 1.02 |
ENSMUST00000174512.4
ENSMUST00000167107.10 |
Zfp101
|
zinc finger protein 101 |
| chr2_+_36120438 | 1.02 |
ENSMUST00000062069.6
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr5_+_139238089 | 1.01 |
ENSMUST00000130326.8
|
Get4
|
golgi to ER traffic protein 4 |
| chr7_-_48106079 | 0.98 |
ENSMUST00000094384.4
|
Mrgprb1
|
MAS-related GPR, member B1 |
| chr17_+_28059099 | 0.95 |
ENSMUST00000233752.2
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr2_+_26899935 | 0.90 |
ENSMUST00000114005.9
ENSMUST00000114004.8 ENSMUST00000114006.8 ENSMUST00000114007.8 ENSMUST00000133807.2 |
Cacfd1
|
calcium channel flower domain containing 1 |
| chr17_-_35191127 | 0.85 |
ENSMUST00000087328.4
|
Hspa1a
|
heat shock protein 1A |
| chr4_+_59003121 | 0.84 |
ENSMUST00000095070.4
ENSMUST00000174664.2 |
Dnajc25
Gm20503
|
DnaJ heat shock protein family (Hsp40) member C25 predicted gene 20503 |
| chr19_-_8691797 | 0.81 |
ENSMUST00000206797.2
|
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr11_-_72098239 | 0.80 |
ENSMUST00000238653.2
ENSMUST00000108505.2 |
4933427D14Rik
|
RIKEN cDNA 4933427D14 gene |
| chr11_-_8614497 | 0.75 |
ENSMUST00000020695.13
|
Tns3
|
tensin 3 |
| chr17_-_37574640 | 0.73 |
ENSMUST00000080759.5
|
Olfr98
|
olfactory receptor 98 |
| chr12_-_83534482 | 0.72 |
ENSMUST00000177959.8
ENSMUST00000178756.8 |
Dpf3
|
D4, zinc and double PHD fingers, family 3 |
| chr18_+_46983105 | 0.71 |
ENSMUST00000025358.4
ENSMUST00000234519.2 |
Lvrn
|
laeverin |
| chr4_-_63779562 | 0.67 |
ENSMUST00000030047.3
|
Tnfsf8
|
tumor necrosis factor (ligand) superfamily, member 8 |
| chr18_+_10617773 | 0.67 |
ENSMUST00000002551.5
ENSMUST00000234207.2 |
Snrpd1
|
small nuclear ribonucleoprotein D1 |
| chr8_+_61085853 | 0.66 |
ENSMUST00000161421.2
|
Mfap3l
|
microfibrillar-associated protein 3-like |
| chr5_-_86780277 | 0.64 |
ENSMUST00000116553.9
|
Tmprss11f
|
transmembrane protease, serine 11f |
| chr11_-_115824290 | 0.63 |
ENSMUST00000021097.10
|
Recql5
|
RecQ protein-like 5 |
| chr6_+_60921456 | 0.62 |
ENSMUST00000129603.4
ENSMUST00000204333.2 |
Mmrn1
|
multimerin 1 |
| chr9_+_109760931 | 0.62 |
ENSMUST00000165876.8
|
Map4
|
microtubule-associated protein 4 |
| chr12_-_40273173 | 0.62 |
ENSMUST00000001672.12
|
Ifrd1
|
interferon-related developmental regulator 1 |
| chr11_+_19874354 | 0.60 |
ENSMUST00000093299.13
|
Spred2
|
sprouty-related EVH1 domain containing 2 |
| chr6_-_106725895 | 0.59 |
ENSMUST00000205004.2
|
Il5ra
|
interleukin 5 receptor, alpha |
| chr19_+_46044972 | 0.58 |
ENSMUST00000111899.8
ENSMUST00000099392.10 ENSMUST00000062322.11 |
Pprc1
|
peroxisome proliferative activated receptor, gamma, coactivator-related 1 |
| chr19_-_8691220 | 0.58 |
ENSMUST00000010239.6
|
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr3_+_89738654 | 0.56 |
ENSMUST00000050401.6
|
She
|
src homology 2 domain-containing transforming protein E |
| chr17_+_28059036 | 0.55 |
ENSMUST00000071006.9
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr7_-_48106037 | 0.54 |
ENSMUST00000188095.2
|
Mrgprb1
|
MAS-related GPR, member B1 |
| chr3_+_20111958 | 0.53 |
ENSMUST00000002502.12
|
Hltf
|
helicase-like transcription factor |
| chr17_-_83821716 | 0.52 |
ENSMUST00000025095.9
ENSMUST00000167741.9 |
Cox7a2l
|
cytochrome c oxidase subunit 7A2 like |
| chr9_+_109760856 | 0.49 |
ENSMUST00000169851.8
|
Map4
|
microtubule-associated protein 4 |
| chr7_+_28416194 | 0.49 |
ENSMUST00000032818.13
|
Fbxo17
|
F-box protein 17 |
| chr19_+_46140942 | 0.48 |
ENSMUST00000026254.14
|
Gbf1
|
golgi-specific brefeldin A-resistance factor 1 |
| chr6_-_106725929 | 0.48 |
ENSMUST00000204659.3
|
Il5ra
|
interleukin 5 receptor, alpha |
| chrX_-_73009933 | 0.47 |
ENSMUST00000114372.3
ENSMUST00000033761.13 |
Hcfc1
|
host cell factor C1 |
| chr3_+_92980844 | 0.44 |
ENSMUST00000195847.2
ENSMUST00000179064.2 |
Tdpoz8
|
TD and POZ domain containing 8 |
| chr8_+_85807369 | 0.43 |
ENSMUST00000079764.14
|
Wdr83os
|
WD repeat domain 83 opposite strand |
| chr7_+_23781311 | 0.43 |
ENSMUST00000207002.2
ENSMUST00000068975.6 ENSMUST00000203854.3 |
Zfp180
|
zinc finger protein 180 |
| chr17_+_37716368 | 0.43 |
ENSMUST00000077008.4
|
Olfr107
|
olfactory receptor 107 |
| chr12_+_111409087 | 0.42 |
ENSMUST00000109792.8
|
Tnfaip2
|
tumor necrosis factor, alpha-induced protein 2 |
| chr7_+_28416270 | 0.39 |
ENSMUST00000108279.9
|
Fbxo17
|
F-box protein 17 |
| chr6_+_29361408 | 0.38 |
ENSMUST00000156163.2
|
Calu
|
calumenin |
| chr11_+_115671523 | 0.37 |
ENSMUST00000239299.2
|
Tmem94
|
transmembrane protein 94 |
| chr10_-_80861239 | 0.35 |
ENSMUST00000055125.5
|
Diras1
|
DIRAS family, GTP-binding RAS-like 1 |
| chr11_+_19874403 | 0.33 |
ENSMUST00000093298.12
|
Spred2
|
sprouty-related EVH1 domain containing 2 |
| chr8_-_100143029 | 0.31 |
ENSMUST00000155527.8
ENSMUST00000142129.8 ENSMUST00000093249.11 ENSMUST00000142475.3 ENSMUST00000128860.8 |
Cdh8
|
cadherin 8 |
| chr7_+_127376267 | 0.30 |
ENSMUST00000144406.8
|
Setd1a
|
SET domain containing 1A |
| chr18_+_89215438 | 0.30 |
ENSMUST00000237110.2
|
Cd226
|
CD226 antigen |
| chr8_-_106723026 | 0.30 |
ENSMUST00000227363.2
ENSMUST00000081998.13 |
Dpep2
|
dipeptidase 2 |
| chrX_-_42363663 | 0.29 |
ENSMUST00000016294.8
|
Tenm1
|
teneurin transmembrane protein 1 |
| chr11_+_72098363 | 0.24 |
ENSMUST00000021158.4
|
Txndc17
|
thioredoxin domain containing 17 |
| chr18_-_32082624 | 0.23 |
ENSMUST00000064016.6
|
Gpr17
|
G protein-coupled receptor 17 |
| chr9_+_118307250 | 0.23 |
ENSMUST00000111763.8
|
Eomes
|
eomesodermin |
| chr8_+_85807566 | 0.21 |
ENSMUST00000140621.2
|
Wdr83os
|
WD repeat domain 83 opposite strand |
| chr7_-_126460820 | 0.19 |
ENSMUST00000129812.2
ENSMUST00000106342.8 |
Ino80e
|
INO80 complex subunit E |
| chr10_+_67371295 | 0.19 |
ENSMUST00000145754.8
ENSMUST00000145936.2 |
Egr2
|
early growth response 2 |
| chr5_+_124250360 | 0.18 |
ENSMUST00000024470.13
ENSMUST00000119269.6 ENSMUST00000196627.5 ENSMUST00000199457.5 ENSMUST00000198505.2 |
Ogfod2
|
2-oxoglutarate and iron-dependent oxygenase domain containing 2 |
| chr9_-_64633865 | 0.17 |
ENSMUST00000168366.2
|
Rab11a
|
RAB11A, member RAS oncogene family |
| chr17_-_35178208 | 0.16 |
ENSMUST00000172753.2
|
Hspa1b
|
heat shock protein 1B |
| chr4_+_140428777 | 0.16 |
ENSMUST00000138808.8
ENSMUST00000038893.6 |
Rcc2
|
regulator of chromosome condensation 2 |
| chr7_+_10421716 | 0.16 |
ENSMUST00000117413.9
ENSMUST00000047809.11 |
Nlrp4b
|
NLR family, pyrin domain containing 4B |
| chr7_+_98092628 | 0.14 |
ENSMUST00000098274.5
|
Gucy2d
|
guanylate cyclase 2d |
| chr12_-_115531211 | 0.13 |
ENSMUST00000103536.4
|
Ighv8-11
|
immunoglobulin heavy variable V8-11 |
| chr14_+_53370136 | 0.12 |
ENSMUST00000181793.3
|
Trav6n-6
|
T cell receptor alpha variable 6N-6 |
| chr4_-_68872585 | 0.11 |
ENSMUST00000030036.6
|
Brinp1
|
bone morphogenic protein/retinoic acid inducible neural specific 1 |
| chr2_+_109848224 | 0.05 |
ENSMUST00000150183.9
|
Ccdc34
|
coiled-coil domain containing 34 |
| chr11_-_59484115 | 0.03 |
ENSMUST00000215626.2
|
Olfr223
|
olfactory receptor 223 |
| chr3_-_89309944 | 0.03 |
ENSMUST00000057431.6
|
Lenep
|
lens epithelial protein |
| chr10_-_80861357 | 0.02 |
ENSMUST00000144640.2
|
Diras1
|
DIRAS family, GTP-binding RAS-like 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.7 | GO:0072312 | metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 1.0 | 3.9 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 0.5 | 4.4 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.4 | 3.0 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.4 | 1.4 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.3 | 2.6 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.3 | 2.3 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.3 | 1.4 | GO:0060356 | leucine import(GO:0060356) |
| 0.2 | 3.7 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.2 | 1.0 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.2 | 1.3 | GO:0030473 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.2 | 0.5 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 | 1.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 0.6 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 8.3 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.5 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.1 | 1.3 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 1.4 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 2.5 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.1 | 2.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.2 | GO:0021664 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.1 | 0.2 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.1 | 0.2 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.1 | 2.8 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 0.3 | GO:0060369 | positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.9 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 3.6 | GO:0090501 | RNA phosphodiester bond hydrolysis(GO:0090501) |
| 0.0 | 1.6 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 3.9 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.0 | 0.7 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 1.1 | GO:0032674 | regulation of interleukin-5 production(GO:0032674) |
| 0.0 | 0.2 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 2.2 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 1.1 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 4.7 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.6 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 0.3 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.5 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 | 0.8 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.5 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 1.9 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.0 | 0.7 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 1.8 | GO:0008584 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 2.7 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.2 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 3.5 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 0.8 | GO:0048286 | lung alveolus development(GO:0048286) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.0 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.3 | 2.3 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.2 | 3.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.2 | 4.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 10.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 2.6 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 4.6 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 3.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.8 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 1.3 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.7 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.8 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 3.6 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.3 | GO:0072686 | mitotic spindle(GO:0072686) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 10.7 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.7 | 2.6 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.6 | 1.9 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.6 | 3.9 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.5 | 3.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.4 | 4.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.4 | 3.6 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.2 | 0.9 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 1.4 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 0.8 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 3.5 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 0.6 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.6 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 1.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.6 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 0.3 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.1 | 1.5 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 9.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 4.7 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 2.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 1.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 1.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.2 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 2.6 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 1.0 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 3.7 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 1.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 7.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 11.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.9 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 2.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 5.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.1 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 3.0 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 3.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 3.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 10.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 4.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 2.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 3.9 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 1.1 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.0 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |