Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
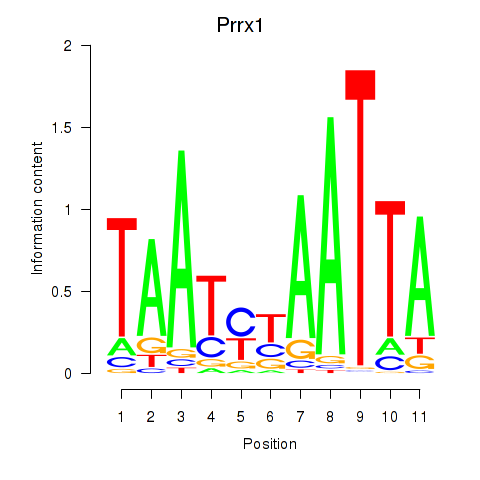
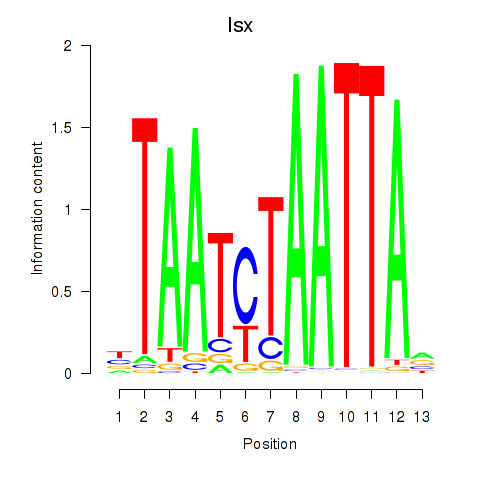
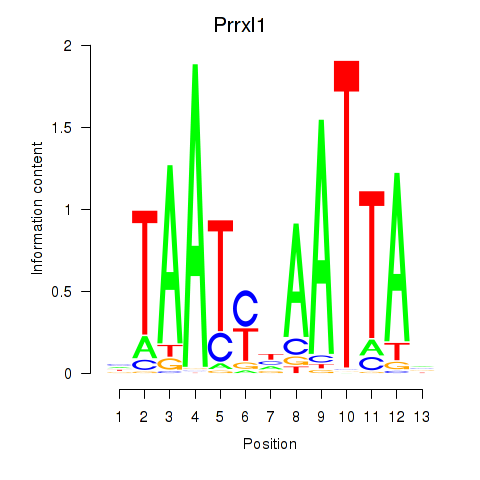
Results for Prrx1_Isx_Prrxl1
Z-value: 1.17
Transcription factors associated with Prrx1_Isx_Prrxl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Prrx1
|
ENSMUSG00000026586.17 | Prrx1 |
|
Isx
|
ENSMUSG00000031621.10 | Isx |
|
Prrxl1
|
ENSMUSG00000041730.15 | Prrxl1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Isx | mm39_v1_chr8_+_75599801_75599855 | 0.72 | 7.9e-13 | Click! |
| Prrx1 | mm39_v1_chr1_-_163141230_163141246 | 0.25 | 3.3e-02 | Click! |
| Prrxl1 | mm39_v1_chr14_+_32321824_32321915 | -0.07 | 5.4e-01 | Click! |
Activity profile of Prrx1_Isx_Prrxl1 motif
Sorted Z-values of Prrx1_Isx_Prrxl1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Prrx1_Isx_Prrxl1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_22055402 | 20.61 |
ENSMUST00000084040.3
|
Defa37
|
defensin, alpha, 37 |
| chr8_+_21917427 | 20.51 |
ENSMUST00000095424.6
|
Defa36
|
defensin, alpha, 36 |
| chr8_+_21787455 | 19.81 |
ENSMUST00000098892.5
|
Defa5
|
defensin, alpha, 5 |
| chr11_+_43046476 | 19.37 |
ENSMUST00000238415.2
|
Atp10b
|
ATPase, class V, type 10B |
| chr8_+_22155813 | 19.17 |
ENSMUST00000075268.5
|
Defa34
|
defensin, alpha, 34 |
| chr8_+_21691577 | 16.76 |
ENSMUST00000110754.2
|
Defa41
|
defensin, alpha, 41 |
| chr8_+_21555054 | 14.55 |
ENSMUST00000078121.4
|
Defa35
|
defensin, alpha, 35 |
| chr8_+_21515561 | 14.23 |
ENSMUST00000076754.3
|
Defa21
|
defensin, alpha, 21 |
| chr6_-_68609426 | 13.19 |
ENSMUST00000103328.3
|
Igkv10-96
|
immunoglobulin kappa variable 10-96 |
| chr3_-_72875187 | 12.34 |
ENSMUST00000167334.8
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr6_+_40619913 | 11.40 |
ENSMUST00000238599.2
|
Mgam
|
maltase-glucoamylase |
| chr6_-_115569504 | 10.61 |
ENSMUST00000112957.2
|
Mkrn2os
|
makorin, ring finger protein 2, opposite strand |
| chr5_-_87682972 | 9.35 |
ENSMUST00000120150.2
|
Sult1b1
|
sulfotransferase family 1B, member 1 |
| chr14_+_80237691 | 8.41 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr11_-_12362136 | 8.27 |
ENSMUST00000174874.8
|
Cobl
|
cordon-bleu WH2 repeat |
| chr5_-_99091681 | 6.98 |
ENSMUST00000162619.8
ENSMUST00000162147.6 |
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr6_+_78347636 | 6.32 |
ENSMUST00000204873.3
|
Reg3b
|
regenerating islet-derived 3 beta |
| chr1_+_93789068 | 6.32 |
ENSMUST00000094663.4
|
Gal3st2
|
galactose-3-O-sulfotransferase 2 |
| chr6_+_78347844 | 6.00 |
ENSMUST00000096904.6
ENSMUST00000203266.2 |
Reg3b
|
regenerating islet-derived 3 beta |
| chr6_+_40763875 | 5.72 |
ENSMUST00000195870.3
|
Mgam2-ps
|
maltase-glucoamylase 2, pseudogene |
| chr6_-_69282389 | 5.61 |
ENSMUST00000103350.3
|
Igkv4-68
|
immunoglobulin kappa variable 4-68 |
| chr1_+_130754413 | 5.48 |
ENSMUST00000027675.14
ENSMUST00000133792.8 |
Pigr
|
polymeric immunoglobulin receptor |
| chr8_+_21999274 | 5.15 |
ENSMUST00000084042.4
|
Defa20
|
defensin, alpha, 20 |
| chr3_+_122688721 | 5.01 |
ENSMUST00000023820.6
|
Fabp2
|
fatty acid binding protein 2, intestinal |
| chr9_-_105973975 | 4.95 |
ENSMUST00000121963.3
|
Col6a4
|
collagen, type VI, alpha 4 |
| chr5_+_64250268 | 4.38 |
ENSMUST00000087324.7
|
Pgm2
|
phosphoglucomutase 2 |
| chr19_+_38384428 | 4.22 |
ENSMUST00000054098.4
|
Slc35g1
|
solute carrier family 35, member G1 |
| chrX_+_111404963 | 4.19 |
ENSMUST00000026602.9
ENSMUST00000113412.3 |
2010106E10Rik
|
RIKEN cDNA 2010106E10 gene |
| chr3_+_103739877 | 4.10 |
ENSMUST00000062945.12
|
Bcl2l15
|
BCLl2-like 15 |
| chr4_-_87724533 | 4.09 |
ENSMUST00000126353.8
ENSMUST00000149357.8 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr3_+_122213420 | 4.06 |
ENSMUST00000029766.9
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr2_-_60552980 | 4.02 |
ENSMUST00000028348.9
ENSMUST00000112517.8 |
Itgb6
|
integrin beta 6 |
| chr19_-_24178000 | 3.91 |
ENSMUST00000233658.3
|
Tjp2
|
tight junction protein 2 |
| chr11_-_94673526 | 3.83 |
ENSMUST00000100554.8
|
Tmem92
|
transmembrane protein 92 |
| chr13_+_4283729 | 3.81 |
ENSMUST00000081326.7
|
Akr1c19
|
aldo-keto reductase family 1, member C19 |
| chr8_-_58106027 | 3.71 |
ENSMUST00000110316.3
|
Galnt7
|
polypeptide N-acetylgalactosaminyltransferase 7 |
| chr10_+_82695055 | 3.54 |
ENSMUST00000218694.2
|
Txnrd1
|
thioredoxin reductase 1 |
| chr12_-_113928438 | 3.52 |
ENSMUST00000103478.4
|
Ighv3-1
|
immunoglobulin heavy variable 3-1 |
| chr15_-_37459570 | 3.43 |
ENSMUST00000119730.8
ENSMUST00000120746.8 |
Ncald
|
neurocalcin delta |
| chr6_-_113411718 | 3.34 |
ENSMUST00000113091.8
|
Cidec
|
cell death-inducing DFFA-like effector c |
| chr7_-_79497940 | 3.32 |
ENSMUST00000107392.8
|
Anpep
|
alanyl (membrane) aminopeptidase |
| chr7_-_79497922 | 3.24 |
ENSMUST00000205502.2
|
Anpep
|
alanyl (membrane) aminopeptidase |
| chrX_-_93256244 | 3.21 |
ENSMUST00000113922.2
|
Eif2s3x
|
eukaryotic translation initiation factor 2, subunit 3, structural gene X-linked |
| chr8_-_58106057 | 3.17 |
ENSMUST00000034021.12
|
Galnt7
|
polypeptide N-acetylgalactosaminyltransferase 7 |
| chr1_-_144052997 | 3.14 |
ENSMUST00000111941.2
ENSMUST00000052375.8 |
Rgs13
|
regulator of G-protein signaling 13 |
| chr7_+_19725390 | 3.10 |
ENSMUST00000207805.2
|
Nlrp9b
|
NLR family, pyrin domain containing 9B |
| chr2_+_155593030 | 3.10 |
ENSMUST00000029140.12
ENSMUST00000132608.2 |
Procr
|
protein C receptor, endothelial |
| chr15_+_37233280 | 3.07 |
ENSMUST00000161405.8
ENSMUST00000022895.15 ENSMUST00000161532.2 |
Grhl2
|
grainyhead like transcription factor 2 |
| chr9_-_96774719 | 2.95 |
ENSMUST00000154146.8
|
Pxylp1
|
2-phosphoxylose phosphatase 1 |
| chr11_+_100978103 | 2.91 |
ENSMUST00000107302.8
ENSMUST00000107303.10 ENSMUST00000017945.15 ENSMUST00000149597.2 |
Mlx
|
MAX-like protein X |
| chr4_-_94538329 | 2.87 |
ENSMUST00000107101.2
|
Lrrc19
|
leucine rich repeat containing 19 |
| chr17_+_34573760 | 2.82 |
ENSMUST00000178562.2
ENSMUST00000025198.15 |
Btnl2
|
butyrophilin-like 2 |
| chr2_+_15531281 | 2.77 |
ENSMUST00000146205.3
|
Malrd1
|
MAM and LDL receptor class A domain containing 1 |
| chr7_+_126575510 | 2.76 |
ENSMUST00000206780.2
|
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr10_+_82695013 | 2.73 |
ENSMUST00000020484.9
ENSMUST00000219962.3 |
Txnrd1
|
thioredoxin reductase 1 |
| chr10_+_115979787 | 2.72 |
ENSMUST00000105271.9
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr1_-_158183894 | 2.67 |
ENSMUST00000004133.11
|
Brinp2
|
bone morphogenic protein/retinoic acid inducible neural-specific 2 |
| chr3_+_159545309 | 2.61 |
ENSMUST00000068952.10
ENSMUST00000198878.2 |
Wls
|
wntless WNT ligand secretion mediator |
| chr18_+_44237474 | 2.57 |
ENSMUST00000081271.7
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr1_+_13738967 | 2.42 |
ENSMUST00000088542.4
|
Xkr9
|
X-linked Kx blood group related 9 |
| chr11_-_117764258 | 2.38 |
ENSMUST00000033230.8
|
Tha1
|
threonine aldolase 1 |
| chr2_-_84481101 | 2.32 |
ENSMUST00000111691.2
|
Ctnnd1
|
catenin (cadherin associated protein), delta 1 |
| chr9_+_78137927 | 2.27 |
ENSMUST00000098537.4
|
Gsta1
|
glutathione S-transferase, alpha 1 (Ya) |
| chr18_+_44237577 | 2.26 |
ENSMUST00000239465.2
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr10_-_75946790 | 2.26 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr4_-_87724512 | 2.25 |
ENSMUST00000148059.2
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr17_-_78991691 | 2.18 |
ENSMUST00000145480.2
|
Strn
|
striatin, calmodulin binding protein |
| chr8_-_41469786 | 2.16 |
ENSMUST00000117735.8
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr3_+_89622323 | 2.13 |
ENSMUST00000098924.9
|
Adar
|
adenosine deaminase, RNA-specific |
| chr7_+_99808452 | 2.11 |
ENSMUST00000032967.4
|
Lipt2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chr11_+_58311921 | 2.10 |
ENSMUST00000013797.3
|
1810065E05Rik
|
RIKEN cDNA 1810065E05 gene |
| chr6_-_69800923 | 2.10 |
ENSMUST00000103368.3
|
Igkv5-43
|
immunoglobulin kappa chain variable 5-43 |
| chr16_-_45352346 | 2.07 |
ENSMUST00000232600.2
|
Gm17783
|
predicted gene, 17783 |
| chr6_+_41511248 | 2.04 |
ENSMUST00000192366.2
ENSMUST00000103286.2 |
Trbj1-3
|
T cell receptor beta joining 1-3 |
| chr17_-_37178079 | 2.03 |
ENSMUST00000025329.13
ENSMUST00000174195.8 |
Trim15
|
tripartite motif-containing 15 |
| chr12_-_114355789 | 2.03 |
ENSMUST00000103486.2
|
Ighv6-3
|
immunoglobulin heavy variable 6-3 |
| chr1_+_78286946 | 2.01 |
ENSMUST00000036172.10
|
Sgpp2
|
sphingosine-1-phosphate phosphatase 2 |
| chr18_+_37453427 | 2.01 |
ENSMUST00000078271.4
|
Pcdhb5
|
protocadherin beta 5 |
| chr7_+_99808526 | 1.96 |
ENSMUST00000207825.2
|
Lipt2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chr15_+_65682066 | 1.94 |
ENSMUST00000211878.2
|
Efr3a
|
EFR3 homolog A |
| chr1_-_174749379 | 1.94 |
ENSMUST00000055294.4
|
Grem2
|
gremlin 2, DAN family BMP antagonist |
| chr7_+_126575752 | 1.91 |
ENSMUST00000206346.2
|
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr18_+_31742565 | 1.90 |
ENSMUST00000164667.2
|
B930094E09Rik
|
RIKEN cDNA B930094E09 gene |
| chr11_+_94827050 | 1.90 |
ENSMUST00000001547.8
|
Col1a1
|
collagen, type I, alpha 1 |
| chr11_+_21041291 | 1.90 |
ENSMUST00000093290.12
|
Peli1
|
pellino 1 |
| chr2_+_152578164 | 1.88 |
ENSMUST00000038368.9
ENSMUST00000109824.2 |
Id1
|
inhibitor of DNA binding 1, HLH protein |
| chr10_+_24025188 | 1.87 |
ENSMUST00000020174.7
ENSMUST00000220041.2 |
Stx7
|
syntaxin 7 |
| chr9_+_78164402 | 1.84 |
ENSMUST00000217203.2
|
Gm3776
|
predicted gene 3776 |
| chr4_-_129121676 | 1.84 |
ENSMUST00000106051.8
|
C77080
|
expressed sequence C77080 |
| chr10_+_69761597 | 1.82 |
ENSMUST00000182269.8
ENSMUST00000183261.8 ENSMUST00000183074.8 |
Ank3
|
ankyrin 3, epithelial |
| chr11_-_30218167 | 1.81 |
ENSMUST00000006629.14
|
Sptbn1
|
spectrin beta, non-erythrocytic 1 |
| chr9_-_39515420 | 1.79 |
ENSMUST00000042485.11
ENSMUST00000141370.8 |
AW551984
|
expressed sequence AW551984 |
| chr9_+_119170486 | 1.79 |
ENSMUST00000175743.8
ENSMUST00000176397.8 |
Acaa1a
|
acetyl-Coenzyme A acyltransferase 1A |
| chrX_-_99638466 | 1.74 |
ENSMUST00000053373.2
|
P2ry4
|
pyrimidinergic receptor P2Y, G-protein coupled, 4 |
| chr10_+_69761784 | 1.73 |
ENSMUST00000181974.8
ENSMUST00000182795.8 ENSMUST00000182437.8 |
Ank3
|
ankyrin 3, epithelial |
| chr8_+_22073022 | 1.71 |
ENSMUST00000098887.4
|
Defa28
|
defensin, alpha, 28 |
| chr3_+_57332735 | 1.70 |
ENSMUST00000029377.8
|
Tm4sf4
|
transmembrane 4 superfamily member 4 |
| chrM_+_7758 | 1.69 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chrX_+_138511360 | 1.68 |
ENSMUST00000113026.2
|
Rnf128
|
ring finger protein 128 |
| chr5_+_110088292 | 1.64 |
ENSMUST00000170826.2
|
Gm15446
|
predicted gene 15446 |
| chr4_-_94538370 | 1.64 |
ENSMUST00000053419.9
|
Lrrc19
|
leucine rich repeat containing 19 |
| chr17_+_36353844 | 1.64 |
ENSMUST00000097331.2
|
Gm6034
|
predicted gene 6034 |
| chr11_-_99313078 | 1.59 |
ENSMUST00000017741.4
|
Krt12
|
keratin 12 |
| chr3_+_60380463 | 1.58 |
ENSMUST00000195077.6
ENSMUST00000193647.6 ENSMUST00000195001.2 ENSMUST00000192807.6 |
Mbnl1
|
muscleblind like splicing factor 1 |
| chr6_+_36364990 | 1.56 |
ENSMUST00000172278.8
|
Chrm2
|
cholinergic receptor, muscarinic 2, cardiac |
| chr14_+_73475335 | 1.56 |
ENSMUST00000044405.8
|
Lpar6
|
lysophosphatidic acid receptor 6 |
| chr17_+_71326510 | 1.56 |
ENSMUST00000073211.13
ENSMUST00000024847.14 |
Myom1
|
myomesin 1 |
| chr4_+_103000248 | 1.54 |
ENSMUST00000106855.2
|
Mier1
|
MEIR1 treanscription regulator |
| chrX_-_133012457 | 1.52 |
ENSMUST00000159259.3
ENSMUST00000113275.10 |
Nox1
|
NADPH oxidase 1 |
| chr9_+_80072361 | 1.51 |
ENSMUST00000184480.8
|
Myo6
|
myosin VI |
| chr12_-_113896002 | 1.51 |
ENSMUST00000103463.3
|
Ighv14-1
|
immunoglobulin heavy variable 14-1 |
| chr10_+_101994841 | 1.49 |
ENSMUST00000020039.13
|
Mgat4c
|
MGAT4 family, member C |
| chr3_-_79053182 | 1.47 |
ENSMUST00000118340.7
|
Rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chrM_+_11735 | 1.47 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr7_-_46316767 | 1.46 |
ENSMUST00000168335.3
ENSMUST00000107669.9 |
Tph1
|
tryptophan hydroxylase 1 |
| chr10_+_69761314 | 1.45 |
ENSMUST00000182692.8
ENSMUST00000092433.12 |
Ank3
|
ankyrin 3, epithelial |
| chr18_+_74575623 | 1.42 |
ENSMUST00000121875.8
|
Myo5b
|
myosin VB |
| chrX_+_10118544 | 1.41 |
ENSMUST00000049910.13
|
Otc
|
ornithine transcarbamylase |
| chr4_+_3940747 | 1.40 |
ENSMUST00000119403.2
|
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr7_+_126575781 | 1.38 |
ENSMUST00000206450.2
ENSMUST00000205830.2 |
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr18_-_39620115 | 1.38 |
ENSMUST00000097592.9
ENSMUST00000115571.8 |
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr17_+_71326542 | 1.37 |
ENSMUST00000179759.3
|
Myom1
|
myomesin 1 |
| chr13_-_3968157 | 1.37 |
ENSMUST00000223258.2
ENSMUST00000091853.12 |
Net1
|
neuroepithelial cell transforming gene 1 |
| chr4_-_110149916 | 1.37 |
ENSMUST00000106601.8
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr4_-_58499398 | 1.37 |
ENSMUST00000107570.2
|
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr18_+_74575567 | 1.34 |
ENSMUST00000074157.13
|
Myo5b
|
myosin VB |
| chrM_+_7779 | 1.33 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr18_+_39126325 | 1.33 |
ENSMUST00000137497.9
|
Arhgap26
|
Rho GTPase activating protein 26 |
| chr12_-_84664001 | 1.33 |
ENSMUST00000221070.2
ENSMUST00000021666.6 ENSMUST00000223107.2 |
Abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr12_+_80691275 | 1.32 |
ENSMUST00000217889.2
|
Slc39a9
|
solute carrier family 39 (zinc transporter), member 9 |
| chr7_-_25112256 | 1.29 |
ENSMUST00000200880.4
ENSMUST00000074040.4 |
Cxcl17
|
chemokine (C-X-C motif) ligand 17 |
| chr13_+_110063364 | 1.29 |
ENSMUST00000117420.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr14_-_110992533 | 1.27 |
ENSMUST00000078386.4
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr5_+_104350475 | 1.26 |
ENSMUST00000066708.7
|
Dmp1
|
dentin matrix protein 1 |
| chr9_+_35334878 | 1.22 |
ENSMUST00000154652.8
|
Cdon
|
cell adhesion molecule-related/down-regulated by oncogenes |
| chr9_+_21634779 | 1.22 |
ENSMUST00000034713.9
|
Ldlr
|
low density lipoprotein receptor |
| chr6_-_37419030 | 1.22 |
ENSMUST00000041093.6
|
Creb3l2
|
cAMP responsive element binding protein 3-like 2 |
| chr9_+_119170360 | 1.21 |
ENSMUST00000039784.12
|
Acaa1a
|
acetyl-Coenzyme A acyltransferase 1A |
| chr5_-_67973195 | 1.20 |
ENSMUST00000141443.2
|
Atp8a1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 |
| chr12_-_57592907 | 1.19 |
ENSMUST00000044380.8
|
Foxa1
|
forkhead box A1 |
| chr10_+_69761630 | 1.18 |
ENSMUST00000182029.8
|
Ank3
|
ankyrin 3, epithelial |
| chrM_+_10167 | 1.18 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr3_-_116505469 | 1.18 |
ENSMUST00000153108.6
|
Slc35a3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member 3 |
| chr11_-_54140462 | 1.18 |
ENSMUST00000019060.6
|
Csf2
|
colony stimulating factor 2 (granulocyte-macrophage) |
| chr15_-_37008011 | 1.18 |
ENSMUST00000226671.2
|
Zfp706
|
zinc finger protein 706 |
| chr13_+_49761506 | 1.16 |
ENSMUST00000021822.7
|
Ogn
|
osteoglycin |
| chr9_-_71803354 | 1.15 |
ENSMUST00000184448.8
|
Tcf12
|
transcription factor 12 |
| chr11_+_55104609 | 1.15 |
ENSMUST00000108867.2
|
Slc36a1
|
solute carrier family 36 (proton/amino acid symporter), member 1 |
| chr5_-_5564730 | 1.13 |
ENSMUST00000115445.8
ENSMUST00000179804.8 ENSMUST00000125110.2 ENSMUST00000115446.8 |
Cldn12
|
claudin 12 |
| chr10_+_69369632 | 1.13 |
ENSMUST00000182155.8
ENSMUST00000183169.8 ENSMUST00000183148.8 |
Ank3
|
ankyrin 3, epithelial |
| chr16_-_16418397 | 1.12 |
ENSMUST00000159542.8
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr5_+_117378510 | 1.07 |
ENSMUST00000111975.3
|
Taok3
|
TAO kinase 3 |
| chr3_-_49711706 | 1.06 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chr7_-_102408573 | 1.06 |
ENSMUST00000210453.3
ENSMUST00000232246.3 ENSMUST00000239110.2 ENSMUST00000060187.15 ENSMUST00000168007.3 |
Olfr560
Olfr78
|
olfactory receptor 560 olfactory receptor 78 |
| chr3_+_84500854 | 1.05 |
ENSMUST00000062623.4
|
Tigd4
|
tigger transposable element derived 4 |
| chr9_+_117888124 | 1.05 |
ENSMUST00000123690.2
|
Azi2
|
5-azacytidine induced gene 2 |
| chrX_+_10118600 | 1.05 |
ENSMUST00000115528.3
|
Otc
|
ornithine transcarbamylase |
| chr3_+_60380243 | 1.04 |
ENSMUST00000195724.6
|
Mbnl1
|
muscleblind like splicing factor 1 |
| chr1_-_136877277 | 1.04 |
ENSMUST00000168126.7
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr18_-_47466378 | 1.03 |
ENSMUST00000126684.2
ENSMUST00000156422.8 |
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr1_+_83094481 | 1.03 |
ENSMUST00000027351.13
ENSMUST00000113437.9 ENSMUST00000186832.2 |
Ccl20
|
chemokine (C-C motif) ligand 20 |
| chr13_+_49658249 | 1.03 |
ENSMUST00000051504.8
|
Ecm2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chrM_+_9870 | 1.02 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr17_+_3447465 | 1.02 |
ENSMUST00000072156.7
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr2_-_89774457 | 1.02 |
ENSMUST00000090695.3
|
Olfr1259
|
olfactory receptor 1259 |
| chr13_+_19526322 | 1.01 |
ENSMUST00000184430.2
|
Trgj4
|
T cell receptor gamma joining 4 |
| chr18_+_39126178 | 0.97 |
ENSMUST00000097593.9
|
Arhgap26
|
Rho GTPase activating protein 26 |
| chr10_+_80971054 | 0.96 |
ENSMUST00000125261.2
|
Zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr10_-_76073656 | 0.95 |
ENSMUST00000099572.10
ENSMUST00000020452.12 ENSMUST00000099571.10 |
Prmt2
|
protein arginine N-methyltransferase 2 |
| chr7_+_79939747 | 0.95 |
ENSMUST00000205864.2
|
Vps33b
|
vacuolar protein sorting 33B |
| chr9_-_57065572 | 0.93 |
ENSMUST00000065358.9
|
Commd4
|
COMM domain containing 4 |
| chr6_-_83098255 | 0.93 |
ENSMUST00000205023.2
ENSMUST00000146328.4 ENSMUST00000151393.7 ENSMUST00000032111.11 ENSMUST00000113936.10 |
Wbp1
|
WW domain binding protein 1 |
| chr7_-_143239600 | 0.92 |
ENSMUST00000208017.2
ENSMUST00000152703.2 |
Tnfrsf23
|
tumor necrosis factor receptor superfamily, member 23 |
| chr6_+_34722887 | 0.91 |
ENSMUST00000123823.8
ENSMUST00000136907.8 |
Cald1
|
caldesmon 1 |
| chr11_+_51989508 | 0.90 |
ENSMUST00000020608.3
|
Ppp2ca
|
protein phosphatase 2 (formerly 2A), catalytic subunit, alpha isoform |
| chr5_+_52940391 | 0.90 |
ENSMUST00000031077.13
ENSMUST00000113904.9 ENSMUST00000199840.2 |
Zcchc4
|
zinc finger, CCHC domain containing 4 |
| chr7_-_110443557 | 0.89 |
ENSMUST00000177462.8
ENSMUST00000176716.3 ENSMUST00000176746.8 ENSMUST00000177236.8 |
Rnf141
|
ring finger protein 141 |
| chr4_-_43710231 | 0.89 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chr10_+_127257077 | 0.89 |
ENSMUST00000168780.8
|
R3hdm2
|
R3H domain containing 2 |
| chr15_-_103071720 | 0.88 |
ENSMUST00000231198.2
|
Smug1
|
single-strand selective monofunctional uracil DNA glycosylase |
| chr6_+_122490534 | 0.87 |
ENSMUST00000032210.14
ENSMUST00000148517.8 |
Mfap5
|
microfibrillar associated protein 5 |
| chr9_+_53212871 | 0.87 |
ENSMUST00000051014.2
|
Exph5
|
exophilin 5 |
| chr2_+_20742115 | 0.87 |
ENSMUST00000114606.8
ENSMUST00000114608.3 |
Etl4
|
enhancer trap locus 4 |
| chr12_+_80690985 | 0.87 |
ENSMUST00000219405.2
ENSMUST00000085245.7 |
Slc39a9
|
solute carrier family 39 (zinc transporter), member 9 |
| chr17_+_36432553 | 0.86 |
ENSMUST00000173128.2
|
Gm19684
|
predicted gene, 19684 |
| chr5_-_151113619 | 0.85 |
ENSMUST00000062015.15
ENSMUST00000110483.9 |
Stard13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr16_-_44153498 | 0.84 |
ENSMUST00000047446.13
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr3_+_32490525 | 0.83 |
ENSMUST00000108242.2
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr9_+_105520154 | 0.83 |
ENSMUST00000190358.2
ENSMUST00000191268.7 ENSMUST00000065778.13 ENSMUST00000188784.2 |
Pik3r4
|
phosphoinositide-3-kinase regulatory subunit 4 |
| chrX_+_9751861 | 0.83 |
ENSMUST00000067529.9
ENSMUST00000086165.4 |
Sytl5
|
synaptotagmin-like 5 |
| chr10_+_79832313 | 0.83 |
ENSMUST00000132517.8
|
Abca7
|
ATP-binding cassette, sub-family A (ABC1), member 7 |
| chrX_-_133012600 | 0.83 |
ENSMUST00000033610.13
|
Nox1
|
NADPH oxidase 1 |
| chr7_-_44541787 | 0.81 |
ENSMUST00000208551.2
ENSMUST00000208253.2 ENSMUST00000207654.2 ENSMUST00000207278.2 |
Med25
|
mediator complex subunit 25 |
| chr8_+_75720286 | 0.81 |
ENSMUST00000211863.2
|
Hmgxb4
|
HMG box domain containing 4 |
| chr3_+_124114504 | 0.81 |
ENSMUST00000058994.6
|
Tram1l1
|
translocation associated membrane protein 1-like 1 |
| chr5_-_138617952 | 0.80 |
ENSMUST00000063262.11
ENSMUST00000085852.11 ENSMUST00000110905.9 |
Zfp68
|
zinc finger protein 68 |
| chr2_-_86866875 | 0.80 |
ENSMUST00000215978.3
|
Olfr1105
|
olfactory receptor 1105 |
| chr11_+_31950452 | 0.79 |
ENSMUST00000109409.8
ENSMUST00000020537.9 |
Nsg2
|
neuron specific gene family member 2 |
| chr12_+_119356318 | 0.79 |
ENSMUST00000221866.2
|
Macc1
|
metastasis associated in colon cancer 1 |
| chr9_-_48822399 | 0.78 |
ENSMUST00000003826.8
|
Htr3a
|
5-hydroxytryptamine (serotonin) receptor 3A |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0002414 | immune response in mucosal-associated lymphoid tissue(GO:0002386) immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 1.7 | 7.0 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 1.4 | 4.1 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 1.2 | 6.0 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 1.2 | 8.3 | GO:0001757 | somite specification(GO:0001757) |
| 1.1 | 6.3 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 1.0 | 3.1 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.9 | 12.3 | GO:0036476 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.9 | 6.3 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.9 | 2.6 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.8 | 2.5 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.7 | 4.4 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.7 | 2.1 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.6 | 1.9 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.6 | 4.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.6 | 7.3 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.6 | 3.9 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.5 | 4.6 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.5 | 1.5 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.5 | 2.4 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.5 | 2.3 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.5 | 1.9 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.5 | 2.8 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.5 | 2.8 | GO:0032439 | endosome localization(GO:0032439) |
| 0.5 | 2.7 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.4 | 6.1 | GO:0007379 | segment specification(GO:0007379) |
| 0.4 | 2.0 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.4 | 1.2 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.4 | 2.3 | GO:0060690 | epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.4 | 1.1 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.3 | 1.0 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.3 | 1.4 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.3 | 0.7 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.3 | 4.6 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.3 | 1.0 | GO:0060156 | vascular transport(GO:0010232) milk ejection(GO:0060156) |
| 0.3 | 6.3 | GO:0051923 | sulfation(GO:0051923) |
| 0.3 | 1.6 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.3 | 6.6 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.3 | 1.2 | GO:0010899 | regulation of phosphatidylcholine catabolic process(GO:0010899) positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.3 | 0.8 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 0.3 | 1.7 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.3 | 0.8 | GO:2001178 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.3 | 1.3 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.2 | 7.7 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.2 | 2.9 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) |
| 0.2 | 1.2 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.2 | 1.9 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.2 | 0.7 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.2 | 0.7 | GO:1990428 | miRNA transport(GO:1990428) |
| 0.2 | 1.3 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.2 | 1.9 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.2 | 1.0 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.2 | 0.6 | GO:0071504 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) response to heparin(GO:0071503) cellular response to heparin(GO:0071504) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.2 | 1.4 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.2 | 0.6 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.2 | 1.7 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.2 | 1.5 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.2 | 0.5 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.2 | 0.5 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.2 | 2.9 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 0.9 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.2 | 0.7 | GO:1902530 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.2 | 2.3 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.2 | 1.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 1.5 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.4 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.1 | 0.6 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 1.6 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 1.2 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.1 | 3.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 10.0 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.9 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 1.2 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.1 | 0.5 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.4 | GO:1903632 | positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) |
| 0.1 | 0.4 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.1 | 0.8 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.1 | 0.5 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 1.4 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 0.9 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 1.2 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 0.8 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 1.8 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.1 | 1.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.1 | 5.0 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.1 | 0.3 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.1 | 4.0 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.1 | 4.3 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 1.6 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.1 | 0.5 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 6.9 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.1 | 2.5 | GO:0046519 | sphingoid metabolic process(GO:0046519) |
| 0.1 | 0.4 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 3.7 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 38.6 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.1 | 0.5 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.1 | 0.7 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 0.8 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 1.3 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.1 | 3.2 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 3.6 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 1.3 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.1 | 0.5 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.1 | 1.8 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.1 | 0.5 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.1 | 0.8 | GO:0033007 | negative regulation of mast cell activation involved in immune response(GO:0033007) |
| 0.1 | 0.5 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 2.3 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.1 | 0.4 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 0.3 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.1 | 0.6 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.1 | 0.5 | GO:1903772 | regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.1 | 1.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.7 | GO:0032049 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 0.3 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 0.3 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 0.3 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.1 | 0.4 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 2.1 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 0.2 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.1 | 0.3 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 0.7 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.1 | 0.3 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 1.3 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 | 0.2 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.1 | 0.6 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.7 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 2.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.8 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 4.5 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 0.9 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 2.5 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 | 0.5 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.1 | 1.5 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 | 0.4 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 1.0 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 2.4 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.2 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.0 | 3.2 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.5 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 1.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.4 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 2.5 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.2 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 1.7 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 0.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.9 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.6 | GO:0051382 | kinetochore assembly(GO:0051382) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 0.1 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.0 | 1.1 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 6.9 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.8 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.6 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 0.6 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.3 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.5 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.6 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.3 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 1.5 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 0.2 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.4 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.9 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 1.2 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.3 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 3.3 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.7 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.5 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.5 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 | 2.9 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 2.2 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.2 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.0 | 0.3 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.4 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.2 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.0 | GO:0070100 | regulation of chemokine-mediated signaling pathway(GO:0070099) negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.7 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 1.4 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.4 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.3 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.9 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.5 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.2 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.7 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 0.3 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 1.0 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.2 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.4 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 1.9 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.9 | 8.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.9 | 4.6 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.7 | 7.5 | GO:0045179 | apical cortex(GO:0045179) |
| 0.6 | 1.9 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.5 | 1.6 | GO:1904602 | serotonin-activated cation-selective channel complex(GO:1904602) |
| 0.5 | 6.6 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.4 | 1.2 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.4 | 3.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.4 | 2.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.3 | 12.3 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.3 | 8.4 | GO:0042581 | specific granule(GO:0042581) |
| 0.3 | 1.8 | GO:0032437 | spectrin(GO:0008091) cuticular plate(GO:0032437) |
| 0.3 | 2.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.2 | 1.1 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.2 | 6.0 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.2 | 0.6 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.2 | 1.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.2 | 3.2 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.2 | 2.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.2 | 1.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.6 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.1 | 3.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 5.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 2.1 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.3 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 4.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.5 | GO:0071547 | piP-body(GO:0071547) |
| 0.1 | 1.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 2.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.5 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 4.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 0.8 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.4 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 7.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.8 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.9 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 0.8 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 2.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.2 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 0.9 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.4 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 13.5 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 0.7 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.7 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 0.2 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.5 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.3 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 4.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 3.7 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 1.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 2.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 3.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 1.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.6 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 4.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.4 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 6.8 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.2 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 1.4 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 2.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 35.8 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 2.6 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 25.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.5 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.9 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 1.3 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 1.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.2 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 4.1 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 12.1 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.2 | GO:0030673 | axolemma(GO:0030673) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 23.7 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 2.0 | 6.0 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 1.9 | 9.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 1.6 | 6.3 | GO:0033797 | selenate reductase activity(GO:0033797) |
| 1.2 | 7.0 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 1.0 | 3.0 | GO:0008775 | acetate CoA-transferase activity(GO:0008775) |
| 0.9 | 6.3 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.7 | 4.4 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.7 | 2.1 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.6 | 21.8 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.6 | 2.5 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.6 | 2.4 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.6 | 1.7 | GO:0045030 | UTP-activated nucleotide receptor activity(GO:0045030) |
| 0.6 | 5.0 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.5 | 2.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.5 | 2.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.5 | 1.5 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.5 | 5.5 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.4 | 2.9 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.4 | 2.3 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.4 | 1.6 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.4 | 1.2 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.3 | 1.6 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.3 | 6.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.3 | 1.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.3 | 3.3 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.3 | 6.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 1.4 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.2 | 0.7 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.2 | 1.4 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.2 | 0.9 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.2 | 0.8 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.2 | 1.9 | GO:0036122 | BMP binding(GO:0036122) |
| 0.2 | 2.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 2.6 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.2 | 2.9 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.2 | 8.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 2.9 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.2 | 1.0 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.2 | 1.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 0.7 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.1 | 2.0 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.1 | 2.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.7 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 1.0 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.1 | 1.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.4 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 3.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.5 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.1 | 0.5 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.1 | 0.2 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.1 | 1.5 | GO:0031697 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) beta-1 adrenergic receptor binding(GO:0031697) |
| 0.1 | 1.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.3 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 0.5 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.4 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 0.3 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.3 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.1 | 2.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.9 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 0.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.3 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.1 | 0.4 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 2.3 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.1 | 0.9 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 14.0 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.1 | 1.0 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 1.9 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 6.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.5 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.1 | 0.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.2 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.1 | 3.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 1.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.2 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.1 | 2.3 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 0.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 2.3 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 2.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.6 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.8 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 23.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 6.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.2 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 0.3 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.4 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.5 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.7 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.4 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.2 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.2 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.0 | 0.7 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.5 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 1.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 1.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.5 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.5 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.3 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 2.7 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.6 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 3.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 1.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.7 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 7.9 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 1.0 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.8 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.5 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 1.8 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 2.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.0 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.4 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.5 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 3.9 | GO:0016746 | transferase activity, transferring acyl groups(GO:0016746) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.7 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 0.8 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 4.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 1.9 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 4.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 5.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 5.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 3.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 2.2 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 3.8 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 2.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 2.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.1 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.5 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 2.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.1 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.7 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.4 | ST ADRENERGIC | Adrenergic Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 11.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 1.4 | 19.8 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.7 | 9.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.4 | 20.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.3 | 7.0 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.3 | 4.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 3.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 6.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 3.9 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 7.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.9 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 2.5 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.9 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 2.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 0.5 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 1.9 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.6 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 6.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 0.9 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 2.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.7 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 2.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 6.0 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 1.6 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 1.9 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 4.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.8 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.7 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 2.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.8 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 2.8 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 3.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.1 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 4.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.0 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 2.0 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 2.6 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.7 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |