Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
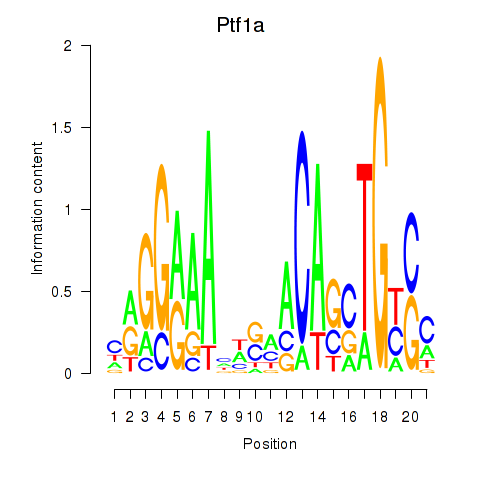
Results for Ptf1a
Z-value: 1.25
Transcription factors associated with Ptf1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ptf1a
|
ENSMUSG00000026735.3 | Ptf1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ptf1a | mm39_v1_chr2_+_19450443_19450474 | 0.29 | 1.4e-02 | Click! |
Activity profile of Ptf1a motif
Sorted Z-values of Ptf1a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Ptf1a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_114104740 | 13.63 |
ENSMUST00000103473.2
|
Ighv9-3
|
immunoglobulin heavy variable V9-3 |
| chr7_+_28240262 | 12.83 |
ENSMUST00000119180.4
|
Sycn
|
syncollin |
| chr6_-_69626340 | 12.49 |
ENSMUST00000198328.2
|
Igkv4-53
|
immunoglobulin kappa variable 4-53 |
| chr6_-_41291634 | 11.17 |
ENSMUST00000064324.12
|
Try5
|
trypsin 5 |
| chr8_-_112417633 | 10.27 |
ENSMUST00000034435.7
|
Ctrb1
|
chymotrypsinogen B1 |
| chr6_+_68233361 | 9.40 |
ENSMUST00000103320.3
|
Igkv14-111
|
immunoglobulin kappa variable 14-111 |
| chr6_-_69584812 | 8.88 |
ENSMUST00000103359.3
|
Igkv4-55
|
immunoglobulin kappa variable 4-55 |
| chr6_+_70549568 | 8.63 |
ENSMUST00000196940.2
ENSMUST00000103397.3 |
Igkv3-10
|
immunoglobulin kappa variable 3-10 |
| chr14_-_63654478 | 7.86 |
ENSMUST00000014597.5
|
Blk
|
B lymphoid kinase |
| chr6_+_41279199 | 7.75 |
ENSMUST00000031913.5
|
Try4
|
trypsin 4 |
| chr6_+_41024260 | 7.62 |
ENSMUST00000103263.3
|
Trbv2
|
T cell receptor beta, variable 2 |
| chr6_-_136758716 | 6.95 |
ENSMUST00000078095.11
ENSMUST00000032338.10 |
Gucy2c
|
guanylate cyclase 2c |
| chr12_-_114263874 | 6.53 |
ENSMUST00000103482.2
ENSMUST00000194159.2 |
Ighv9-4
|
immunoglobulin heavy variable 9-4 |
| chr2_-_34851561 | 6.37 |
ENSMUST00000028234.12
|
Traf1
|
TNF receptor-associated factor 1 |
| chr11_+_66847446 | 6.32 |
ENSMUST00000211300.2
ENSMUST00000150220.2 |
Tmem238l
|
transmembrane protein 238 like |
| chr8_+_123920233 | 6.07 |
ENSMUST00000212773.2
|
Dpep1
|
dipeptidase 1 |
| chr6_-_69394425 | 5.99 |
ENSMUST00000199160.2
|
Igkv4-61
|
immunoglobulin kappa chain variable 4-61 |
| chr4_-_133615075 | 5.94 |
ENSMUST00000003741.16
ENSMUST00000105894.11 |
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr6_-_70318164 | 5.84 |
ENSMUST00000103389.3
|
Igkv8-19
|
immunoglobulin kappa variable 8-19 |
| chr12_-_114057841 | 5.79 |
ENSMUST00000103471.2
ENSMUST00000195884.2 |
Ighv9-1
|
immunoglobulin heavy variable 9-1 |
| chr11_-_99045894 | 5.79 |
ENSMUST00000103134.4
|
Ccr7
|
chemokine (C-C motif) receptor 7 |
| chr2_+_180367056 | 5.78 |
ENSMUST00000094218.4
|
Slc17a9
|
solute carrier family 17, member 9 |
| chr6_+_41118120 | 5.71 |
ENSMUST00000103273.3
|
Trbv15
|
T cell receptor beta, variable 15 |
| chr13_+_19528728 | 5.44 |
ENSMUST00000179181.3
|
Trgc4
|
T cell receptor gamma, constant 4 |
| chr2_+_164611812 | 5.41 |
ENSMUST00000088248.13
ENSMUST00000001439.7 |
Ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr5_-_121025645 | 4.99 |
ENSMUST00000086368.12
|
Oas1g
|
2'-5' oligoadenylate synthetase 1G |
| chr15_-_76501041 | 4.98 |
ENSMUST00000073428.7
|
Slc39a4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr12_-_114073050 | 4.70 |
ENSMUST00000103472.4
|
Ighv9-2
|
immunoglobulin heavy variable V9-2 |
| chr7_+_103978678 | 4.70 |
ENSMUST00000180136.2
ENSMUST00000106847.9 |
Trim34b
|
tripartite motif-containing 34B |
| chr8_+_123920682 | 4.68 |
ENSMUST00000212409.2
|
Dpep1
|
dipeptidase 1 |
| chr14_-_32907023 | 4.65 |
ENSMUST00000130509.10
ENSMUST00000061753.15 |
Wdfy4
|
WD repeat and FYVE domain containing 4 |
| chr6_+_41435846 | 4.53 |
ENSMUST00000031910.8
|
Prss1
|
protease, serine 1 (trypsin 1) |
| chr7_-_130924021 | 4.19 |
ENSMUST00000046611.9
|
Cuzd1
|
CUB and zona pellucida-like domains 1 |
| chr2_+_164790139 | 3.86 |
ENSMUST00000017881.3
|
Mmp9
|
matrix metallopeptidase 9 |
| chr6_-_69162381 | 3.78 |
ENSMUST00000103344.3
|
Igkv4-74
|
immunoglobulin kappa variable 4-74 |
| chr1_-_171434882 | 3.70 |
ENSMUST00000111277.2
ENSMUST00000004827.14 |
Ly9
|
lymphocyte antigen 9 |
| chr12_-_115276219 | 3.66 |
ENSMUST00000103529.4
|
Ighv1-58
|
immunoglobulin heavy variable 1-58 |
| chr7_-_97848688 | 3.58 |
ENSMUST00000098278.4
|
B3gnt6
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 6 (core 3 synthase) |
| chr1_-_171434944 | 3.54 |
ENSMUST00000068878.14
|
Ly9
|
lymphocyte antigen 9 |
| chr7_-_140793990 | 3.48 |
ENSMUST00000026573.7
ENSMUST00000170841.9 |
Lmntd2
|
lamin tail domain containing 2 |
| chr6_+_41369290 | 3.43 |
ENSMUST00000049079.9
|
Gm5771
|
predicted gene 5771 |
| chr8_+_82069177 | 3.36 |
ENSMUST00000213285.2
ENSMUST00000217122.2 ENSMUST00000215332.2 |
Inpp4b
|
inositol polyphosphate-4-phosphatase, type II |
| chr7_-_79492091 | 3.18 |
ENSMUST00000049004.8
|
Anpep
|
alanyl (membrane) aminopeptidase |
| chr1_+_36510670 | 3.17 |
ENSMUST00000153128.2
|
Cnnm4
|
cyclin M4 |
| chr16_-_21980200 | 3.16 |
ENSMUST00000115379.2
|
Igf2bp2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr7_+_97100523 | 3.09 |
ENSMUST00000206658.2
|
Kctd14
|
potassium channel tetramerisation domain containing 14 |
| chr12_-_114451189 | 3.03 |
ENSMUST00000103493.3
|
Ighv1-4
|
immunoglobulin heavy variable 1-4 |
| chr6_+_41331039 | 3.02 |
ENSMUST00000072103.7
|
Try10
|
trypsin 10 |
| chr6_-_115785695 | 3.00 |
ENSMUST00000081840.6
|
Rpl32
|
ribosomal protein L32 |
| chr7_+_118199375 | 2.90 |
ENSMUST00000121744.9
|
Tmc5
|
transmembrane channel-like gene family 5 |
| chr19_-_7218363 | 2.85 |
ENSMUST00000236769.2
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chrX_-_161747552 | 2.79 |
ENSMUST00000038769.3
|
S100g
|
S100 calcium binding protein G |
| chr14_-_32907446 | 2.75 |
ENSMUST00000159606.2
|
Wdfy4
|
WD repeat and FYVE domain containing 4 |
| chr10_+_60182630 | 2.73 |
ENSMUST00000020301.14
ENSMUST00000105460.8 ENSMUST00000170507.8 |
Vsir
|
V-set immunoregulatory receptor |
| chr6_-_68784692 | 2.72 |
ENSMUST00000103334.4
|
Igkv4-90
|
immunoglobulin kappa chain variable 4-90 |
| chr3_-_83749036 | 2.69 |
ENSMUST00000029623.11
|
Tlr2
|
toll-like receptor 2 |
| chr9_+_20927271 | 2.63 |
ENSMUST00000086399.6
|
Icam1
|
intercellular adhesion molecule 1 |
| chr14_-_51295099 | 2.33 |
ENSMUST00000227764.2
|
Rnase12
|
ribonuclease, RNase A family, 12 (non-active) |
| chr11_+_48691175 | 2.33 |
ENSMUST00000020640.8
|
Rack1
|
receptor for activated C kinase 1 |
| chr4_-_63965161 | 2.32 |
ENSMUST00000107377.10
|
Tnc
|
tenascin C |
| chr11_-_74480870 | 2.19 |
ENSMUST00000145524.2
ENSMUST00000102521.9 |
Rap1gap2
|
RAP1 GTPase activating protein 2 |
| chr5_-_99400698 | 2.15 |
ENSMUST00000031276.15
ENSMUST00000168092.8 |
Rasgef1b
|
RasGEF domain family, member 1B |
| chr15_-_77283286 | 2.13 |
ENSMUST00000175919.8
ENSMUST00000176074.9 |
Apol7a
|
apolipoprotein L 7a |
| chr10_+_94350687 | 2.07 |
ENSMUST00000065060.12
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr9_+_59563872 | 2.05 |
ENSMUST00000215623.2
ENSMUST00000215660.2 ENSMUST00000217353.2 |
Pkm
|
pyruvate kinase, muscle |
| chr17_+_8144822 | 2.02 |
ENSMUST00000036370.8
|
Tagap
|
T cell activation Rho GTPase activating protein |
| chr14_-_21898992 | 2.01 |
ENSMUST00000124549.9
|
Comtd1
|
catechol-O-methyltransferase domain containing 1 |
| chr9_+_59563838 | 1.92 |
ENSMUST00000163694.4
|
Pkm
|
pyruvate kinase, muscle |
| chr10_+_75047897 | 1.91 |
ENSMUST00000218766.2
|
Specc1l
|
sperm antigen with calponin homology and coiled-coil domains 1-like |
| chr9_+_59614877 | 1.82 |
ENSMUST00000128944.8
ENSMUST00000098661.10 |
Gramd2
|
GRAM domain containing 2 |
| chr6_+_65567373 | 1.77 |
ENSMUST00000114236.2
|
Tnip3
|
TNFAIP3 interacting protein 3 |
| chr19_-_7218512 | 1.70 |
ENSMUST00000025675.11
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr19_-_9876745 | 1.68 |
ENSMUST00000237725.2
|
Incenp
|
inner centromere protein |
| chr6_-_69261303 | 1.66 |
ENSMUST00000103349.2
|
Igkv4-69
|
immunoglobulin kappa variable 4-69 |
| chr9_+_108731884 | 1.63 |
ENSMUST00000239562.1
|
Slc26a6
|
solute carrier family 26, member 6 |
| chr11_+_120653613 | 1.62 |
ENSMUST00000105046.4
|
Hmga1b
|
high mobility group AT-hook 1B |
| chr12_+_71062733 | 1.57 |
ENSMUST00000046305.12
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr16_-_44153288 | 1.45 |
ENSMUST00000136381.8
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr11_-_6394352 | 1.32 |
ENSMUST00000093346.6
|
H2az2
|
H2A.Z histone variant 2 |
| chr11_+_101215889 | 1.24 |
ENSMUST00000041095.14
ENSMUST00000107264.2 |
Aoc2
|
amine oxidase, copper containing 2 (retina-specific) |
| chr19_+_16413126 | 1.21 |
ENSMUST00000025602.4
|
Gna14
|
guanine nucleotide binding protein, alpha 14 |
| chr18_-_80484066 | 1.20 |
ENSMUST00000161003.2
|
Ctdp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
| chr17_+_21910767 | 1.18 |
ENSMUST00000072133.5
|
Gm10226
|
predicted gene 10226 |
| chr13_-_61012185 | 1.17 |
ENSMUST00000091569.7
|
4930486L24Rik
|
RIKEN cDNA 4930486L24 gene |
| chr11_+_54582248 | 1.12 |
ENSMUST00000136494.2
|
Rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr2_-_91780731 | 1.08 |
ENSMUST00000111303.2
|
Dgkz
|
diacylglycerol kinase zeta |
| chr3_+_107008867 | 1.06 |
ENSMUST00000038695.6
|
Kcna2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr12_+_112727089 | 1.06 |
ENSMUST00000063888.5
|
Pld4
|
phospholipase D family, member 4 |
| chr14_+_52860023 | 1.04 |
ENSMUST00000103570.2
|
Trav5-1
|
T cell receptor alpha variable 5-1 |
| chr11_-_97242842 | 1.03 |
ENSMUST00000093942.5
|
Gpr179
|
G protein-coupled receptor 179 |
| chr17_+_29899420 | 1.01 |
ENSMUST00000130052.9
|
Cmtr1
|
cap methyltransferase 1 |
| chr7_-_144136194 | 1.01 |
ENSMUST00000033394.8
|
Fadd
|
Fas (TNFRSF6)-associated via death domain |
| chr18_-_80484003 | 0.99 |
ENSMUST00000160434.3
|
Ctdp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
| chr11_+_40624763 | 0.98 |
ENSMUST00000127382.2
|
Nudcd2
|
NudC domain containing 2 |
| chr11_+_98337655 | 0.97 |
ENSMUST00000019456.5
|
Grb7
|
growth factor receptor bound protein 7 |
| chr16_-_63684477 | 0.94 |
ENSMUST00000232654.2
ENSMUST00000064405.8 |
Epha3
|
Eph receptor A3 |
| chr7_+_79939747 | 0.92 |
ENSMUST00000205864.2
|
Vps33b
|
vacuolar protein sorting 33B |
| chr11_+_40624466 | 0.92 |
ENSMUST00000020578.11
|
Nudcd2
|
NudC domain containing 2 |
| chr7_+_18865610 | 0.91 |
ENSMUST00000165913.2
|
Fbxo46
|
F-box protein 46 |
| chr4_-_88357112 | 0.79 |
ENSMUST00000132493.8
ENSMUST00000030221.3 ENSMUST00000151280.8 |
Hacd4
|
3-hydroxyacyl-CoA dehydratase 4 |
| chr5_-_66211842 | 0.77 |
ENSMUST00000200852.4
|
Rbm47
|
RNA binding motif protein 47 |
| chrX_+_159173680 | 0.76 |
ENSMUST00000112408.9
ENSMUST00000112402.8 ENSMUST00000112401.8 ENSMUST00000112400.8 ENSMUST00000112405.9 ENSMUST00000112404.9 ENSMUST00000146805.8 |
Adgrg2
|
adhesion G protein-coupled receptor G2 |
| chr11_-_6394385 | 0.76 |
ENSMUST00000109737.9
|
H2az2
|
H2A.Z histone variant 2 |
| chr11_-_118246566 | 0.74 |
ENSMUST00000155707.3
|
Timp2
|
tissue inhibitor of metalloproteinase 2 |
| chr6_-_69220672 | 0.74 |
ENSMUST00000196201.2
|
Igkv4-71
|
immunoglobulin kappa chain variable 4-71 |
| chr8_+_72662437 | 0.70 |
ENSMUST00000110002.8
ENSMUST00000125802.8 ENSMUST00000131544.8 |
Zfp882
Zfp617
|
zinc finger protein 882 zinc finger protein 617 |
| chr7_-_142628298 | 0.67 |
ENSMUST00000148715.8
|
Trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr2_+_22232314 | 0.62 |
ENSMUST00000044749.14
|
Myo3a
|
myosin IIIA |
| chr5_+_34683141 | 0.61 |
ENSMUST00000125817.8
ENSMUST00000067638.14 |
Sh3bp2
|
SH3-domain binding protein 2 |
| chr5_+_143450329 | 0.59 |
ENSMUST00000045593.12
|
Daglb
|
diacylglycerol lipase, beta |
| chr11_-_73348284 | 0.59 |
ENSMUST00000121209.3
ENSMUST00000127789.3 |
Olfr380
|
olfactory receptor 380 |
| chr4_+_114945905 | 0.58 |
ENSMUST00000171877.8
ENSMUST00000177647.8 ENSMUST00000106548.9 ENSMUST00000030488.3 |
Pdzk1ip1
|
PDZK1 interacting protein 1 |
| chr11_-_118246332 | 0.57 |
ENSMUST00000017610.10
|
Timp2
|
tissue inhibitor of metalloproteinase 2 |
| chr2_-_125624754 | 0.53 |
ENSMUST00000053699.13
|
Secisbp2l
|
SECIS binding protein 2-like |
| chr19_+_40648182 | 0.52 |
ENSMUST00000112231.9
ENSMUST00000127828.8 |
Entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr16_-_63684425 | 0.51 |
ENSMUST00000232049.2
|
Epha3
|
Eph receptor A3 |
| chr9_-_85631361 | 0.49 |
ENSMUST00000039213.15
|
Ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr4_-_36056726 | 0.49 |
ENSMUST00000108124.4
|
Lingo2
|
leucine rich repeat and Ig domain containing 2 |
| chr12_+_117497478 | 0.48 |
ENSMUST00000220781.2
|
Rapgef5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr14_-_8123295 | 0.45 |
ENSMUST00000225775.2
ENSMUST00000224877.2 ENSMUST00000090591.4 |
Il3ra
|
interleukin 3 receptor, alpha chain |
| chr10_+_42554888 | 0.44 |
ENSMUST00000040718.6
|
Ostm1
|
osteopetrosis associated transmembrane protein 1 |
| chr10_-_75768302 | 0.44 |
ENSMUST00000120281.8
ENSMUST00000000924.13 |
Mmp11
|
matrix metallopeptidase 11 |
| chr7_-_4535403 | 0.41 |
ENSMUST00000094897.5
|
Dnaaf3
|
dynein, axonemal assembly factor 3 |
| chr9_-_112016834 | 0.40 |
ENSMUST00000111872.9
ENSMUST00000164754.9 |
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chr19_+_32463151 | 0.37 |
ENSMUST00000025827.10
|
Minpp1
|
multiple inositol polyphosphate histidine phosphatase 1 |
| chr6_-_5256225 | 0.35 |
ENSMUST00000125686.8
|
Pon3
|
paraoxonase 3 |
| chr6_-_68968278 | 0.35 |
ENSMUST00000197966.2
|
Igkv4-81
|
immunoglobulin kappa variable 4-81 |
| chr11_+_97206542 | 0.34 |
ENSMUST00000019026.10
ENSMUST00000132168.2 |
Mrpl45
|
mitochondrial ribosomal protein L45 |
| chrX_-_162859429 | 0.32 |
ENSMUST00000134272.2
|
Siah1b
|
siah E3 ubiquitin protein ligase 1B |
| chr2_+_153742294 | 0.24 |
ENSMUST00000088955.12
ENSMUST00000135501.3 |
Bpifb6
|
BPI fold containing family B, member 6 |
| chr4_-_43706566 | 0.23 |
ENSMUST00000055401.2
|
Olfr71
|
olfactory receptor 71 |
| chr11_+_72332167 | 0.20 |
ENSMUST00000045633.6
|
Mybbp1a
|
MYB binding protein (P160) 1a |
| chr11_-_102076028 | 0.18 |
ENSMUST00000107156.9
ENSMUST00000021297.6 |
Lsm12
|
LSM12 homolog |
| chr11_+_98489117 | 0.15 |
ENSMUST00000142268.3
|
Lrrc3c
|
leucine rich repeat containing 3C |
| chr1_+_151074199 | 0.09 |
ENSMUST00000225273.2
|
Gm47995
|
predicted gene, 47995 |
| chr9_-_106353571 | 0.07 |
ENSMUST00000123555.8
ENSMUST00000125850.2 |
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr11_-_70213348 | 0.07 |
ENSMUST00000019051.3
|
Alox12e
|
arachidonate lipoxygenase, epidermal |
| chr10_-_126737185 | 0.05 |
ENSMUST00000168520.3
ENSMUST00000026504.13 |
Atp23
|
ATP23 metallopeptidase and ATP synthase assembly factor homolog |
| chr17_+_25527616 | 0.04 |
ENSMUST00000015267.5
|
Prss28
|
protease, serine 28 |
| chr11_+_49138278 | 0.04 |
ENSMUST00000109194.2
|
Mgat1
|
mannoside acetylglucosaminyltransferase 1 |
| chr2_-_93787383 | 0.04 |
ENSMUST00000148314.3
|
Gm13889
|
predicted gene 13889 |
| chr11_+_101221431 | 0.01 |
ENSMUST00000103105.10
|
Aoc3
|
amine oxidase, copper containing 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.8 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 1.9 | 5.8 | GO:0097021 | regulation of tolerance induction to self antigen(GO:0002649) lymphocyte migration into lymphoid organs(GO:0097021) positive regulation of thymocyte migration(GO:2000412) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 1.3 | 3.9 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 1.0 | 5.0 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.9 | 2.7 | GO:0042495 | positive regulation of interleukin-18 production(GO:0032741) detection of triacyl bacterial lipopeptide(GO:0042495) response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 0.7 | 5.4 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.7 | 5.9 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.6 | 2.3 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.6 | 2.3 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.5 | 1.6 | GO:0046724 | oxalic acid secretion(GO:0046724) |
| 0.5 | 1.4 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.5 | 2.7 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.5 | 7.2 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.4 | 4.0 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.4 | 2.6 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.4 | 1.3 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.3 | 1.0 | GO:2000451 | positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) |
| 0.2 | 37.3 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.2 | 37.9 | GO:0007586 | digestion(GO:0007586) |
| 0.2 | 1.0 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.2 | 2.2 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.2 | 51.1 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.2 | 3.2 | GO:0070166 | magnesium ion transport(GO:0015693) enamel mineralization(GO:0070166) |
| 0.2 | 1.1 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.2 | 1.6 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.2 | 4.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 3.4 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.2 | 3.2 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.1 | 1.5 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 0.9 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 6.9 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.1 | 0.4 | GO:0038156 | interleukin-3-mediated signaling pathway(GO:0038156) |
| 0.1 | 1.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 7.9 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 1.8 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.1 | 0.4 | GO:0046226 | coumarin catabolic process(GO:0046226) |
| 0.1 | 0.5 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.1 | 0.8 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 3.0 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 0.5 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 1.1 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 1.9 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 1.0 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.6 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 | 6.4 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 1.1 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.8 | GO:0042761 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 1.0 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 4.2 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 0.6 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 3.2 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.4 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.4 | GO:0070286 | motile cilium assembly(GO:0044458) axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.0 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.9 | 2.7 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.8 | 2.3 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.3 | 37.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.3 | 6.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 4.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.2 | 5.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 12.8 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 1.7 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 1.0 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 1.2 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.1 | 2.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.9 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 0.2 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 1.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 1.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 2.2 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 1.0 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 2.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 9.1 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 3.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 8.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.6 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 64.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 5.4 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 4.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.9 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 1.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 10.8 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.9 | 2.7 | GO:0042497 | triacyl lipopeptide binding(GO:0042497) |
| 0.8 | 3.4 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.8 | 4.0 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.7 | 2.2 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 0.5 | 4.6 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.4 | 6.4 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.4 | 1.3 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.4 | 5.0 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.4 | 5.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.4 | 2.8 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.3 | 2.8 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.3 | 37.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.3 | 1.6 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.3 | 6.9 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 1.5 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.2 | 2.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 41.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 1.0 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.2 | 1.0 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.2 | 0.8 | GO:0102344 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.2 | 3.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.4 | GO:0004912 | interleukin-3 receptor activity(GO:0004912) interleukin-3 binding(GO:0019978) |
| 0.1 | 1.4 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 1.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 3.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 5.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 5.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 5.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 3.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.4 | GO:0052743 | inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 0.1 | 7.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.4 | GO:0004063 | aryldialkylphosphatase activity(GO:0004063) |
| 0.1 | 8.9 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 3.6 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.1 | 1.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 1.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.5 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 0.5 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 1.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 1.1 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 1.6 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 1.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 1.8 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 1.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 2.3 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 2.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 3.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.0 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 1.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 14.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 5.9 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 7.4 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.1 | 2.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 3.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.0 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.6 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 20.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.3 | 2.7 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.3 | 5.4 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.2 | 5.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 3.4 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 7.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 5.9 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 5.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 2.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 1.0 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 1.0 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 3.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.3 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 2.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 3.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |