Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
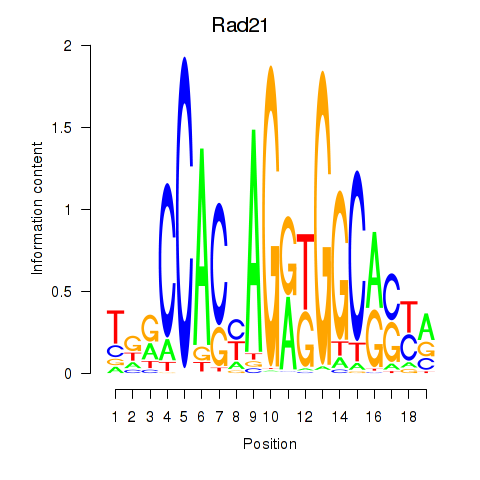
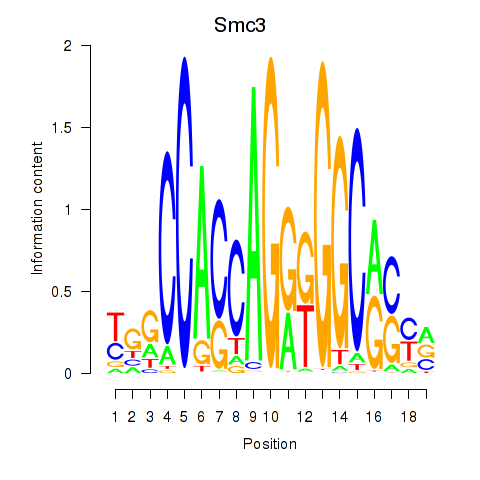
Results for Rad21_Smc3
Z-value: 1.21
Transcription factors associated with Rad21_Smc3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rad21
|
ENSMUSG00000022314.11 | Rad21 |
|
Smc3
|
ENSMUSG00000024974.11 | Smc3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Smc3 | mm39_v1_chr19_+_53588808_53588869 | 0.47 | 3.1e-05 | Click! |
| Rad21 | mm39_v1_chr15_-_51855073_51855159 | 0.45 | 8.0e-05 | Click! |
Activity profile of Rad21_Smc3 motif
Sorted Z-values of Rad21_Smc3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Rad21_Smc3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_106088798 | 9.21 |
ENSMUST00000216850.2
|
Twf2
|
twinfilin actin binding protein 2 |
| chr14_+_53903370 | 8.75 |
ENSMUST00000181768.3
|
Trav3-3
|
T cell receptor alpha variable 3-3 |
| chr4_-_129467430 | 8.28 |
ENSMUST00000102596.8
|
Lck
|
lymphocyte protein tyrosine kinase |
| chr1_-_85664246 | 8.21 |
ENSMUST00000064788.14
|
A630001G21Rik
|
RIKEN cDNA A630001G21 gene |
| chr8_+_106412905 | 8.14 |
ENSMUST00000213019.2
|
Carmil2
|
capping protein regulator and myosin 1 linker 2 |
| chr14_+_53828314 | 7.79 |
ENSMUST00000103654.3
|
Trav9-2
|
T cell receptor alpha variable 9-2 |
| chr14_+_53698556 | 7.09 |
ENSMUST00000181728.3
|
Trav7-4
|
T cell receptor alpha variable 7-4 |
| chr14_+_53725502 | 7.08 |
ENSMUST00000103581.6
|
Trav9-1
|
T cell receptor alpha variable 9-1 |
| chr16_-_10603389 | 7.05 |
ENSMUST00000229866.2
ENSMUST00000038099.6 |
Socs1
|
suppressor of cytokine signaling 1 |
| chr10_-_77879414 | 6.85 |
ENSMUST00000145975.8
ENSMUST00000130972.8 ENSMUST00000128241.8 ENSMUST00000155021.8 ENSMUST00000140636.8 ENSMUST00000148469.8 ENSMUST00000019257.15 ENSMUST00000105395.9 ENSMUST00000156417.8 ENSMUST00000105396.9 ENSMUST00000154374.2 |
Aire
|
autoimmune regulator (autoimmune polyendocrinopathy candidiasis ectodermal dystrophy) |
| chr14_+_53468890 | 6.63 |
ENSMUST00000177705.3
|
Trav9n-3
|
T cell receptor alpha variable 9N-3 |
| chr15_-_75620060 | 6.60 |
ENSMUST00000062002.6
|
Mafa
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein A (avian) |
| chr2_-_117173190 | 6.58 |
ENSMUST00000173541.8
ENSMUST00000172901.8 ENSMUST00000173252.2 |
Rasgrp1
|
RAS guanyl releasing protein 1 |
| chr6_+_41090484 | 6.45 |
ENSMUST00000103267.3
|
Trbv12-1
|
T cell receptor beta, variable 12-1 |
| chr2_+_29509704 | 6.43 |
ENSMUST00000095087.11
ENSMUST00000091146.12 ENSMUST00000102872.11 ENSMUST00000147755.10 |
Rapgef1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr7_+_25386418 | 6.13 |
ENSMUST00000002678.10
|
Tgfb1
|
transforming growth factor, beta 1 |
| chr5_+_137639538 | 6.10 |
ENSMUST00000177466.8
ENSMUST00000166099.3 |
Sap25
|
sin3 associated polypeptide |
| chr14_+_53859114 | 5.95 |
ENSMUST00000103657.6
|
Trav12-3
|
T cell receptor alpha variable 12-3 |
| chr11_+_68322945 | 5.92 |
ENSMUST00000021283.8
|
Pik3r5
|
phosphoinositide-3-kinase regulatory subunit 5 |
| chr3_-_95049655 | 5.81 |
ENSMUST00000013851.4
|
Tnfaip8l2
|
tumor necrosis factor, alpha-induced protein 8-like 2 |
| chr14_+_53853772 | 5.67 |
ENSMUST00000180972.3
|
Trav12-2
|
T cell receptor alpha variable 12-2 |
| chr14_+_53478202 | 5.65 |
ENSMUST00000179583.3
|
Trav12n-3
|
T cell receptor alpha variable 12N-3 |
| chr19_+_42024439 | 5.63 |
ENSMUST00000238137.2
ENSMUST00000026172.3 |
Ankrd2
|
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr14_+_53209905 | 5.55 |
ENSMUST00000196023.2
|
Trav3d-3
|
T cell receptor alpha variable 3D-3 |
| chr14_+_53583346 | 5.44 |
ENSMUST00000103632.4
|
Trav8n-2
|
T cell receptor alpha variable 8n-2 |
| chr14_+_53007210 | 5.42 |
ENSMUST00000178768.4
|
Trav7d-4
|
T cell receptor alpha variable 7D-4 |
| chr14_+_53157900 | 5.40 |
ENSMUST00000178252.3
|
Trav9d-3
|
T cell receptor alpha variable 9D-3 |
| chr14_+_53521042 | 5.39 |
ENSMUST00000197557.2
|
Trav3n-3
|
T cell receptor alpha variable 3N-3 |
| chr14_+_53913598 | 5.36 |
ENSMUST00000103662.6
|
Trav9-4
|
T cell receptor alpha variable 9-4 |
| chr14_+_53680701 | 5.19 |
ENSMUST00000177622.4
|
Trav7-3
|
T cell receptor alpha variable 7-3 |
| chr14_+_53531798 | 5.16 |
ENSMUST00000103626.3
|
Trav9n-4
|
T cell receptor alpha variable 9N-4 |
| chr6_-_71376277 | 5.10 |
ENSMUST00000149415.2
|
Rmnd5a
|
required for meiotic nuclear division 5 homolog A |
| chr14_+_53628092 | 5.10 |
ENSMUST00000103636.4
|
Trav7-2
|
T cell receptor alpha variable 7-2 |
| chr14_+_52860023 | 5.07 |
ENSMUST00000103570.2
|
Trav5-1
|
T cell receptor alpha variable 5-1 |
| chr14_+_53743184 | 4.92 |
ENSMUST00000103583.5
|
Trav10
|
T cell receptor alpha variable 10 |
| chr12_-_84923252 | 4.90 |
ENSMUST00000163189.8
ENSMUST00000110254.9 ENSMUST00000002073.13 |
Ltbp2
|
latent transforming growth factor beta binding protein 2 |
| chr14_+_54119038 | 4.69 |
ENSMUST00000103676.9
ENSMUST00000187163.2 |
Trdv1
|
T cell receptor delta variable 1 |
| chr9_-_44145280 | 4.66 |
ENSMUST00000205968.2
ENSMUST00000206147.2 ENSMUST00000037644.8 |
Cbl
|
Casitas B-lineage lymphoma |
| chr14_+_53220658 | 4.63 |
ENSMUST00000200548.2
|
Trav9d-4
|
T cell receptor alpha variable 9D-4 |
| chr14_+_53775648 | 4.61 |
ENSMUST00000200115.2
ENSMUST00000103650.3 |
Trav12-1
|
T cell receptor alpha variable 12-1 |
| chr14_+_54043954 | 4.56 |
ENSMUST00000103672.9
ENSMUST00000186545.2 |
Trav17
|
T cell receptor alpha variable 17 |
| chr14_+_53015904 | 4.54 |
ENSMUST00000103580.4
|
Trav8d-1
|
T cell receptor alpha variable 8D-1 |
| chr14_+_54014387 | 4.53 |
ENSMUST00000103670.4
|
Trav3-4
|
T cell receptor alpha variable 3-4 |
| chr14_+_53061814 | 4.52 |
ENSMUST00000103648.4
|
Trav11d
|
T cell receptor alpha variable 11D |
| chr14_+_53556255 | 4.49 |
ENSMUST00000179607.3
|
Trav6n-7
|
T cell receptor alpha variable 6N-7 |
| chr1_+_135060431 | 4.42 |
ENSMUST00000187985.7
ENSMUST00000049449.11 |
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr14_+_53153256 | 4.34 |
ENSMUST00000197007.2
ENSMUST00000103593.3 |
Trav12d-2
|
T cell receptor alpha variable 12D-2 |
| chr17_+_34354556 | 4.32 |
ENSMUST00000236322.2
ENSMUST00000237402.2 |
H2-DMa
|
histocompatibility 2, class II, locus DMa |
| chr14_+_53029773 | 4.31 |
ENSMUST00000178426.4
|
Trav9d-1
|
T cell receptor alpha variable 9D-1 |
| chr9_-_44145309 | 4.25 |
ENSMUST00000206720.2
|
Cbl
|
Casitas B-lineage lymphoma |
| chr14_+_53954133 | 4.24 |
ENSMUST00000103641.6
|
Trav7-6
|
T cell receptor alpha variable 7-6 |
| chr4_-_126362372 | 4.19 |
ENSMUST00000097888.10
ENSMUST00000239399.2 |
Ago1
|
argonaute RISC catalytic subunit 1 |
| chr5_-_136596299 | 4.16 |
ENSMUST00000004097.16
|
Cux1
|
cut-like homeobox 1 |
| chr18_-_77802145 | 4.13 |
ENSMUST00000237203.2
|
8030462N17Rik
|
RIKEN cDNA 8030462N17 gene |
| chr14_+_53562089 | 4.08 |
ENSMUST00000178100.3
|
Trav7n-6
|
T cell receptor alpha variable 7N-6 |
| chr17_+_34423054 | 3.96 |
ENSMUST00000138491.2
|
Tap2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr14_+_52892115 | 3.95 |
ENSMUST00000198019.2
|
Trav7-1
|
T cell receptor alpha variable 7-1 |
| chr14_+_52921421 | 3.93 |
ENSMUST00000200127.2
|
Trav7d-2
|
T cell receptor alpha variable 7D-2 |
| chr14_+_53210216 | 3.91 |
ENSMUST00000103599.3
|
Trav3d-3
|
T cell receptor alpha variable 3D-3 |
| chr8_+_112262729 | 3.91 |
ENSMUST00000172856.8
|
Znrf1
|
zinc and ring finger 1 |
| chr16_+_48251600 | 3.86 |
ENSMUST00000023330.8
|
Morc1
|
microrchidia 1 |
| chr14_+_53666165 | 3.85 |
ENSMUST00000180549.3
|
Trav6-3
|
T cell receptor alpha variable 6-3 |
| chr14_+_53239170 | 3.80 |
ENSMUST00000179701.2
|
Trav5d-4
|
T cell receptor alpha variable 5D-4 |
| chr14_+_53392969 | 3.67 |
ENSMUST00000198682.2
ENSMUST00000103587.4 |
Trav12n-1
|
T cell receptor alpha variable 12N-1 |
| chr19_+_7394951 | 3.67 |
ENSMUST00000159348.3
|
2700081O15Rik
|
RIKEN cDNA 2700081O15 gene |
| chr9_-_57552844 | 3.65 |
ENSMUST00000216979.2
ENSMUST00000034863.8 |
Csk
|
c-src tyrosine kinase |
| chr8_-_13304154 | 3.65 |
ENSMUST00000204916.3
ENSMUST00000033825.11 |
Adprhl1
|
ADP-ribosylhydrolase like 1 |
| chr19_+_6450553 | 3.55 |
ENSMUST00000146831.8
ENSMUST00000035716.15 ENSMUST00000138555.8 ENSMUST00000167240.8 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr14_+_53384376 | 3.55 |
ENSMUST00000199753.2
|
Trav7n-5
|
T cell receptor alpha variable 7N-5 |
| chr14_+_53521353 | 3.54 |
ENSMUST00000103625.3
|
Trav3n-3
|
T cell receptor alpha variable 3N-3 |
| chr15_+_74593041 | 3.45 |
ENSMUST00000070923.3
|
Them6
|
thioesterase superfamily member 6 |
| chr17_-_56343625 | 3.41 |
ENSMUST00000003268.11
|
Sh3gl1
|
SH3-domain GRB2-like 1 |
| chr7_+_16044336 | 3.41 |
ENSMUST00000136781.2
|
Bbc3
|
BCL2 binding component 3 |
| chr18_+_34910064 | 3.36 |
ENSMUST00000043775.9
ENSMUST00000224715.2 |
Kdm3b
|
KDM3B lysine (K)-specific demethylase 3B |
| chr17_-_56343531 | 3.33 |
ENSMUST00000233803.2
|
Sh3gl1
|
SH3-domain GRB2-like 1 |
| chr12_-_100995305 | 3.32 |
ENSMUST00000068411.5
|
Ccdc88c
|
coiled-coil domain containing 88C |
| chr18_-_49888606 | 3.30 |
ENSMUST00000025383.10
ENSMUST00000163590.9 |
Dtwd2
|
DTW domain containing 2 |
| chr12_-_100995242 | 3.28 |
ENSMUST00000085096.10
|
Ccdc88c
|
coiled-coil domain containing 88C |
| chr7_-_44785480 | 3.26 |
ENSMUST00000211246.2
ENSMUST00000210197.2 |
Flt3l
|
FMS-like tyrosine kinase 3 ligand |
| chr4_-_132073048 | 3.25 |
ENSMUST00000084250.11
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr5_-_137739863 | 3.22 |
ENSMUST00000061789.14
|
Nyap1
|
neuronal tyrosine-phosphorylated phosphoinositide 3-kinase adaptor 1 |
| chr14_+_52990824 | 3.22 |
ENSMUST00000180717.3
ENSMUST00000183820.2 |
Trav6d-4
|
T cell receptor alpha variable 6D-4 |
| chr4_-_132072988 | 3.20 |
ENSMUST00000030726.13
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr17_-_34109513 | 3.19 |
ENSMUST00000173386.2
ENSMUST00000114361.9 ENSMUST00000173492.9 |
Kifc1
|
kinesin family member C1 |
| chr14_+_53081922 | 3.18 |
ENSMUST00000181360.3
ENSMUST00000183652.2 |
Trav12d-1
|
T cell receptor alpha variable 12D-1 |
| chr19_+_5928649 | 3.16 |
ENSMUST00000136833.8
ENSMUST00000141362.2 |
Slc25a45
|
solute carrier family 25, member 45 |
| chr2_+_172821620 | 3.16 |
ENSMUST00000109125.8
ENSMUST00000109126.5 ENSMUST00000050442.15 |
Spo11
|
SPO11 initiator of meiotic double stranded breaks |
| chr14_+_53707213 | 3.13 |
ENSMUST00000103643.4
|
Trav8-1
|
T cell receptor alpha variable 8-1 |
| chr16_-_20245071 | 3.06 |
ENSMUST00000115547.9
ENSMUST00000096199.5 |
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr14_+_53167246 | 3.03 |
ENSMUST00000177703.3
|
Trav12d-3
|
T cell receptor alpha variable 12D-3 |
| chr5_-_137739364 | 3.00 |
ENSMUST00000149512.3
|
Nyap1
|
neuronal tyrosine-phosphorylated phosphoinositide 3-kinase adaptor 1 |
| chr5_+_137517140 | 2.99 |
ENSMUST00000031727.10
|
Gigyf1
|
GRB10 interacting GYF protein 1 |
| chr2_-_91793847 | 2.96 |
ENSMUST00000028667.10
|
Dgkz
|
diacylglycerol kinase zeta |
| chr16_-_20245138 | 2.95 |
ENSMUST00000079158.13
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr1_+_52669849 | 2.92 |
ENSMUST00000165859.8
|
Nemp2
|
nuclear envelope integral membrane protein 2 |
| chr14_+_52818032 | 2.90 |
ENSMUST00000103569.3
|
Trav3-1
|
T cell receptor alpha variable 3-1 |
| chr14_+_53048391 | 2.90 |
ENSMUST00000103646.5
|
Trav10d
|
T cell receptor alpha variable 10D |
| chr14_+_52875909 | 2.87 |
ENSMUST00000103571.2
|
Trav6-1
|
T cell receptor alpha variable 6-1 |
| chr17_+_27136065 | 2.85 |
ENSMUST00000078961.6
|
Kifc5b
|
kinesin family member C5B |
| chr14_+_53550361 | 2.83 |
ENSMUST00000179997.2
|
Trav5n-4
|
T cell receptor alpha variable 5N-4 |
| chr7_+_16043502 | 2.82 |
ENSMUST00000002152.13
|
Bbc3
|
BCL2 binding component 3 |
| chr7_-_126616472 | 2.78 |
ENSMUST00000206416.2
ENSMUST00000162069.3 ENSMUST00000200948.2 ENSMUST00000202798.2 |
Pagr1a
Gm42742
|
PAXIP1 associated glutamate rich protein 1A predicted gene 42742 |
| chr8_-_13304068 | 2.78 |
ENSMUST00000168498.8
|
Adprhl1
|
ADP-ribosylhydrolase like 1 |
| chr14_+_53941464 | 2.75 |
ENSMUST00000103664.6
|
Trav5-4
|
T cell receptor alpha variable 5-4 |
| chr14_+_53279810 | 2.72 |
ENSMUST00000198439.5
ENSMUST00000103605.3 |
Trav8d-2
|
T cell receptor alpha variable 8D-2 |
| chr14_+_53440579 | 2.69 |
ENSMUST00000198913.5
ENSMUST00000180097.2 |
Trav9n-2
|
T cell receptor alpha variable 9N-2 |
| chr19_+_6450641 | 2.61 |
ENSMUST00000113467.2
|
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr7_-_92523396 | 2.57 |
ENSMUST00000209074.2
ENSMUST00000208356.2 ENSMUST00000032877.11 |
Ddias
|
DNA damage-induced apoptosis suppressor |
| chr19_-_5538370 | 2.56 |
ENSMUST00000124334.8
|
Mus81
|
MUS81 structure-specific endonuclease subunit |
| chr10_+_17931459 | 2.55 |
ENSMUST00000154718.8
ENSMUST00000126390.8 ENSMUST00000164556.8 ENSMUST00000150029.8 |
Reps1
|
RalBP1 associated Eps domain containing protein |
| chr1_-_38168697 | 2.55 |
ENSMUST00000027251.12
|
Rev1
|
REV1, DNA directed polymerase |
| chr5_-_143301113 | 2.55 |
ENSMUST00000046418.3
|
E130309D02Rik
|
RIKEN cDNA E130309D02 gene |
| chr14_+_55842002 | 2.54 |
ENSMUST00000138037.2
|
Irf9
|
interferon regulatory factor 9 |
| chr12_-_26465253 | 2.54 |
ENSMUST00000020971.14
|
Rnf144a
|
ring finger protein 144A |
| chr19_-_24257904 | 2.52 |
ENSMUST00000081333.11
|
Fxn
|
frataxin |
| chr7_-_44785815 | 2.51 |
ENSMUST00000146760.7
|
Flt3l
|
FMS-like tyrosine kinase 3 ligand |
| chr5_+_144705346 | 2.46 |
ENSMUST00000128550.8
|
Trrap
|
transformation/transcription domain-associated protein |
| chr14_+_53947359 | 2.43 |
ENSMUST00000103638.6
|
Trav6-7-dv9
|
T cell receptor alpha variable 6-7-DV9 |
| chr5_+_150597204 | 2.43 |
ENSMUST00000202170.4
ENSMUST00000016569.11 |
Pds5b
|
PDS5 cohesin associated factor B |
| chrX_-_101232978 | 2.40 |
ENSMUST00000033683.8
|
Rps4x
|
ribosomal protein S4, X-linked |
| chr17_+_27904155 | 2.39 |
ENSMUST00000231669.2
ENSMUST00000097360.3 ENSMUST00000231236.2 |
Pacsin1
|
protein kinase C and casein kinase substrate in neurons 1 |
| chr12_-_26465130 | 2.39 |
ENSMUST00000222082.2
ENSMUST00000062149.6 |
Rnf144a
|
ring finger protein 144A |
| chr2_-_33321306 | 2.37 |
ENSMUST00000113158.8
|
Zbtb34
|
zinc finger and BTB domain containing 34 |
| chr14_+_53328803 | 2.37 |
ENSMUST00000103609.2
|
Trav7n-4
|
T cell receptor alpha variable 7N-4 |
| chr13_+_51254852 | 2.35 |
ENSMUST00000095797.6
|
Spin1
|
spindlin 1 |
| chr16_-_10994135 | 2.32 |
ENSMUST00000037633.16
|
Zc3h7a
|
zinc finger CCCH type containing 7 A |
| chr11_-_45835737 | 2.30 |
ENSMUST00000129820.8
|
Lsm11
|
U7 snRNP-specific Sm-like protein LSM11 |
| chr19_+_46293210 | 2.29 |
ENSMUST00000236591.2
|
Nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 |
| chr18_-_10610048 | 2.28 |
ENSMUST00000115864.8
ENSMUST00000145320.2 ENSMUST00000097670.10 |
Esco1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr5_+_64317550 | 2.28 |
ENSMUST00000101195.9
|
Tbc1d1
|
TBC1 domain family, member 1 |
| chr1_-_133537953 | 2.28 |
ENSMUST00000164574.2
ENSMUST00000166291.8 ENSMUST00000164096.2 ENSMUST00000166915.8 |
Snrpe
|
small nuclear ribonucleoprotein E |
| chr14_+_53257873 | 2.27 |
ENSMUST00000196756.2
|
Trav7d-6
|
T cell receptor alpha variable 7D-6 |
| chr17_-_32074754 | 2.27 |
ENSMUST00000024839.6
|
Sik1
|
salt inducible kinase 1 |
| chr5_+_137516810 | 2.26 |
ENSMUST00000197624.5
|
Gigyf1
|
GRB10 interacting GYF protein 1 |
| chr5_+_120787253 | 2.25 |
ENSMUST00000156722.2
|
Rasal1
|
RAS protein activator like 1 (GAP1 like) |
| chr14_+_44162791 | 2.24 |
ENSMUST00000160298.2
|
Gm8113
|
predicted gene 8113 |
| chr15_+_100659622 | 2.22 |
ENSMUST00000023776.13
|
Slc4a8
|
solute carrier family 4 (anion exchanger), member 8 |
| chr18_-_10610332 | 2.19 |
ENSMUST00000025142.13
|
Esco1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr5_+_115644727 | 2.19 |
ENSMUST00000067268.15
ENSMUST00000086523.7 ENSMUST00000212819.3 |
Pxn
|
paxillin |
| chr2_+_90927053 | 2.19 |
ENSMUST00000132741.3
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr8_+_88999031 | 2.18 |
ENSMUST00000169037.9
|
Adcy7
|
adenylate cyclase 7 |
| chr18_-_77801624 | 2.18 |
ENSMUST00000074653.6
|
8030462N17Rik
|
RIKEN cDNA 8030462N17 gene |
| chr14_+_53129643 | 2.18 |
ENSMUST00000199746.5
ENSMUST00000178824.2 |
Trav9d-2
|
T cell receptor alpha variable 9D-2 |
| chr5_+_150597292 | 2.18 |
ENSMUST00000038900.15
|
Pds5b
|
PDS5 cohesin associated factor B |
| chr6_+_47812584 | 2.17 |
ENSMUST00000079881.11
ENSMUST00000114598.3 |
Zfp398
|
zinc finger protein 398 |
| chr18_-_53877579 | 2.15 |
ENSMUST00000049811.8
ENSMUST00000237062.2 |
Cep120
|
centrosomal protein 120 |
| chr8_-_13304096 | 2.14 |
ENSMUST00000171619.2
|
Adprhl1
|
ADP-ribosylhydrolase like 1 |
| chr16_+_16714333 | 2.11 |
ENSMUST00000027373.12
ENSMUST00000232247.2 |
Ppm1f
|
protein phosphatase 1F (PP2C domain containing) |
| chr7_-_126625657 | 2.11 |
ENSMUST00000205568.2
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr5_+_114845821 | 2.09 |
ENSMUST00000094441.11
|
Tchp
|
trichoplein, keratin filament binding |
| chr7_-_142050663 | 2.08 |
ENSMUST00000238347.2
|
Prr33
|
proline rich 33 |
| chr14_+_53464230 | 2.04 |
ENSMUST00000196674.2
ENSMUST00000103619.3 |
Trav12n-2
|
T cell receptor alpha variable 12N-2 |
| chr7_-_126625617 | 2.03 |
ENSMUST00000032916.6
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr9_+_123307758 | 1.97 |
ENSMUST00000026269.4
|
Limd1
|
LIM domains containing 1 |
| chr17_+_48761916 | 1.97 |
ENSMUST00000074574.13
|
Unc5cl
|
unc-5 family C-terminal like |
| chr1_-_143578542 | 1.96 |
ENSMUST00000018337.9
|
Cdc73
|
cell division cycle 73, Paf1/RNA polymerase II complex component |
| chr3_-_95125002 | 1.96 |
ENSMUST00000107209.8
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chr16_-_20244631 | 1.96 |
ENSMUST00000077867.10
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr7_+_108533613 | 1.96 |
ENSMUST00000033342.7
|
Eif3f
|
eukaryotic translation initiation factor 3, subunit F |
| chr8_+_22682816 | 1.96 |
ENSMUST00000033866.9
|
Vps36
|
vacuolar protein sorting 36 |
| chr7_-_126625739 | 1.95 |
ENSMUST00000205461.2
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr5_+_115373895 | 1.94 |
ENSMUST00000081497.13
|
Pop5
|
processing of precursor 5, ribonuclease P/MRP family (S. cerevisiae) |
| chr5_-_136164840 | 1.92 |
ENSMUST00000150406.2
ENSMUST00000006301.11 |
Lrwd1
|
leucine-rich repeats and WD repeat domain containing 1 |
| chr1_+_150268544 | 1.90 |
ENSMUST00000124973.9
|
Tpr
|
translocated promoter region, nuclear basket protein |
| chr12_+_113120023 | 1.89 |
ENSMUST00000049271.13
|
Tedc1
|
tubulin epsilon and delta complex 1 |
| chr8_+_70735477 | 1.86 |
ENSMUST00000087467.12
ENSMUST00000140212.8 ENSMUST00000110124.9 |
Homer3
|
homer scaffolding protein 3 |
| chr13_+_35925296 | 1.84 |
ENSMUST00000163595.3
|
Cdyl
|
chromodomain protein, Y chromosome-like |
| chr1_-_92576563 | 1.82 |
ENSMUST00000053144.4
|
Otos
|
otospiralin |
| chr12_-_100995102 | 1.81 |
ENSMUST00000223097.2
|
Ccdc88c
|
coiled-coil domain containing 88C |
| chr14_+_53245069 | 1.79 |
ENSMUST00000178650.3
|
Trav6d-7
|
T cell receptor alpha variable 6D-7 |
| chr3_-_105594865 | 1.78 |
ENSMUST00000090680.11
|
Ddx20
|
DEAD box helicase 20 |
| chr19_-_36034740 | 1.75 |
ENSMUST00000164639.8
ENSMUST00000166074.2 ENSMUST00000099505.4 |
Htr7
|
5-hydroxytryptamine (serotonin) receptor 7 |
| chr8_-_123159663 | 1.75 |
ENSMUST00000017604.10
|
Cyba
|
cytochrome b-245, alpha polypeptide |
| chr10_-_126866658 | 1.74 |
ENSMUST00000120547.2
ENSMUST00000152054.8 |
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr19_+_57349112 | 1.74 |
ENSMUST00000036407.6
|
Fam160b1
|
family with sequence similarity 160, member B1 |
| chr5_+_64316771 | 1.74 |
ENSMUST00000043893.13
|
Tbc1d1
|
TBC1 domain family, member 1 |
| chr14_+_45567245 | 1.74 |
ENSMUST00000022380.9
|
Psmc6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr13_-_55684317 | 1.73 |
ENSMUST00000021956.9
ENSMUST00000224765.2 |
Ddx41
|
DEAD box helicase 41 |
| chr11_-_69470139 | 1.71 |
ENSMUST00000048139.12
|
Wrap53
|
WD repeat containing, antisense to Trp53 |
| chr4_-_149221998 | 1.71 |
ENSMUST00000176124.8
ENSMUST00000177408.2 ENSMUST00000105695.2 |
Cenps
|
centromere protein S |
| chr3_-_96501443 | 1.70 |
ENSMUST00000145001.2
ENSMUST00000091924.10 |
Polr3gl
|
polymerase (RNA) III (DNA directed) polypeptide G like |
| chr7_+_103893658 | 1.69 |
ENSMUST00000106849.9
ENSMUST00000060315.12 |
Trim34a
|
tripartite motif-containing 34A |
| chr6_+_120813199 | 1.68 |
ENSMUST00000009256.4
|
Bcl2l13
|
BCL2-like 13 (apoptosis facilitator) |
| chr16_-_90841360 | 1.68 |
ENSMUST00000118522.8
|
Paxbp1
|
PAX3 and PAX7 binding protein 1 |
| chr8_-_123159639 | 1.67 |
ENSMUST00000212600.2
|
Cyba
|
cytochrome b-245, alpha polypeptide |
| chr15_-_96358612 | 1.66 |
ENSMUST00000047835.8
|
Scaf11
|
SR-related CTD-associated factor 11 |
| chr7_+_49428082 | 1.64 |
ENSMUST00000032715.13
|
Prmt3
|
protein arginine N-methyltransferase 3 |
| chr4_-_149222057 | 1.64 |
ENSMUST00000030813.10
|
Cenps
|
centromere protein S |
| chr7_-_28071658 | 1.64 |
ENSMUST00000094644.11
|
Plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr12_+_24758724 | 1.62 |
ENSMUST00000153058.8
|
Rrm2
|
ribonucleotide reductase M2 |
| chr8_-_25592001 | 1.61 |
ENSMUST00000128715.8
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr17_+_56920389 | 1.61 |
ENSMUST00000080492.7
|
Rpl36
|
ribosomal protein L36 |
| chr13_-_8921732 | 1.61 |
ENSMUST00000054251.13
ENSMUST00000176813.8 ENSMUST00000175958.2 |
Wdr37
|
WD repeat domain 37 |
| chr12_+_24758968 | 1.60 |
ENSMUST00000154588.2
|
Rrm2
|
ribonucleotide reductase M2 |
| chr7_-_46360066 | 1.60 |
ENSMUST00000143082.4
|
Saal1
|
serum amyloid A-like 1 |
| chr6_+_59185846 | 1.59 |
ENSMUST00000062626.4
|
Tigd2
|
tigger transposable element derived 2 |
| chr7_-_126859790 | 1.59 |
ENSMUST00000035276.5
|
Dctpp1
|
dCTP pyrophosphatase 1 |
| chrX_-_139501246 | 1.59 |
ENSMUST00000112996.9
|
Tsc22d3
|
TSC22 domain family, member 3 |
| chr18_-_38068456 | 1.58 |
ENSMUST00000080033.7
ENSMUST00000115631.8 ENSMUST00000115634.8 |
Diaph1
|
diaphanous related formin 1 |
| chr12_-_8351954 | 1.58 |
ENSMUST00000220073.2
ENSMUST00000037313.6 |
Gdf7
|
growth differentiation factor 7 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.1 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 2.0 | 6.1 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 1.7 | 6.9 | GO:0002458 | peripheral T cell tolerance induction(GO:0002458) peripheral tolerance induction(GO:0002465) regulation of thymocyte migration(GO:2000410) |
| 1.4 | 4.3 | GO:0002501 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 1.4 | 7.0 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 1.3 | 4.0 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) cytosol to ER transport(GO:0046967) |
| 1.2 | 8.6 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 1.1 | 3.4 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 1.1 | 3.2 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 1.0 | 2.9 | GO:0042262 | DNA protection(GO:0042262) |
| 0.9 | 9.2 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.9 | 4.5 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.9 | 6.2 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.9 | 7.1 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 0.9 | 1.8 | GO:0051385 | response to mineralocorticoid(GO:0051385) |
| 0.9 | 2.6 | GO:1903002 | regulation of lipid transport across blood brain barrier(GO:1903000) positive regulation of lipid transport across blood brain barrier(GO:1903002) |
| 0.9 | 2.6 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.8 | 3.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.8 | 3.2 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.8 | 2.4 | GO:1903632 | positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) |
| 0.7 | 3.7 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.7 | 3.3 | GO:0002266 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.6 | 8.3 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.6 | 2.5 | GO:0019230 | proprioception(GO:0019230) |
| 0.6 | 1.8 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.6 | 6.7 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.6 | 6.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.5 | 0.5 | GO:2000424 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.5 | 3.6 | GO:0090005 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.5 | 1.5 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.5 | 1.5 | GO:0046832 | RNA import into nucleus(GO:0006404) mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.5 | 4.1 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.5 | 2.0 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.5 | 1.9 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.5 | 1.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.5 | 6.6 | GO:1902715 | positive regulation of granulocyte macrophage colony-stimulating factor production(GO:0032725) positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.5 | 2.3 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.5 | 1.8 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.5 | 4.5 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.4 | 2.2 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.4 | 1.3 | GO:1990164 | histone H3-S28 phosphorylation(GO:0043988) histone H2A phosphorylation(GO:1990164) |
| 0.4 | 3.4 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.4 | 1.2 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.4 | 2.9 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.4 | 3.9 | GO:2000143 | negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.4 | 6.4 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.4 | 2.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.3 | 2.8 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.3 | 1.4 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.3 | 1.4 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.3 | 1.0 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.3 | 1.6 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.3 | 8.9 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.3 | 1.6 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.3 | 5.6 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.3 | 1.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.3 | 2.8 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.3 | 0.8 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.3 | 0.8 | GO:0034148 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.3 | 0.8 | GO:0060825 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.3 | 11.1 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.3 | 0.8 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.3 | 0.8 | GO:0045404 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.3 | 1.8 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.3 | 3.4 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.2 | 1.2 | GO:1903677 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.2 | 0.5 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.2 | 61.1 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.2 | 2.3 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.2 | 1.3 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.2 | 1.9 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.2 | 1.1 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.2 | 2.1 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.2 | 2.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 | 1.7 | GO:0015675 | nickel cation transport(GO:0015675) |
| 0.2 | 2.9 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.2 | 1.2 | GO:0034627 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.2 | 2.0 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.2 | 5.5 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.2 | 2.5 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.2 | 2.6 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.2 | 0.5 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.2 | 1.3 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.2 | 1.5 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.2 | 0.7 | GO:1902164 | positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.2 | 2.2 | GO:0046607 | positive regulation of centrosome duplication(GO:0010825) positive regulation of centrosome cycle(GO:0046607) |
| 0.2 | 0.8 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.2 | 0.8 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) |
| 0.2 | 1.3 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.2 | 0.5 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.2 | 1.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.2 | 1.6 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.2 | 3.3 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.2 | 1.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 4.6 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 2.2 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 4.5 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 8.0 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.1 | 3.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 2.4 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 1.1 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.1 | 2.5 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.1 | 0.9 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.1 | 5.8 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 1.6 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 2.4 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 0.5 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 1.1 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.1 | 2.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.1 | 5.9 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 1.7 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.1 | 0.5 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.1 | 2.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 1.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 1.2 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 0.4 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 | 1.3 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 3.0 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 1.0 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 1.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 2.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.9 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 2.1 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 1.3 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 0.9 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.1 | 0.3 | GO:1903969 | interleukin-4-mediated signaling pathway(GO:0035771) negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.1 | 0.6 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 4.1 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 0.3 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 0.2 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 2.4 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 0.6 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.1 | 4.1 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.1 | 0.3 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.6 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.1 | 0.4 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.4 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.1 | 0.2 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 0.5 | GO:0051584 | regulation of dopamine uptake involved in synaptic transmission(GO:0051584) regulation of catecholamine uptake involved in synaptic transmission(GO:0051940) positive regulation of synaptic vesicle endocytosis(GO:1900244) |
| 0.1 | 0.4 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.4 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.1 | 0.6 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 0.4 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.1 | 1.3 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 3.5 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.1 | 1.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.5 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 2.2 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 0.2 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.8 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.1 | 5.1 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 3.9 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 2.3 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 0.6 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.1 | 0.1 | GO:0009227 | nucleotide-sugar catabolic process(GO:0009227) |
| 0.1 | 0.5 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.2 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 0.8 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.1 | 1.5 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.1 | 1.0 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.1 | 0.9 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.1 | 9.3 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.1 | 1.4 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 1.4 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.1 | 2.2 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 1.7 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 0.2 | GO:0009814 | defense response, incompatible interaction(GO:0009814) |
| 0.1 | 2.3 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 0.4 | GO:0061366 | axonogenesis involved in innervation(GO:0060385) behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.3 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 1.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 5.4 | GO:0050868 | negative regulation of T cell activation(GO:0050868) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.2 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.7 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 2.7 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 1.7 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 1.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.5 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.6 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 2.7 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0071110 | protein biotinylation(GO:0009305) response to biotin(GO:0070781) histone biotinylation(GO:0071110) |
| 0.0 | 0.6 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.6 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 1.4 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 2.5 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 1.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 1.4 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.0 | 0.6 | GO:0070266 | necroptotic process(GO:0070266) |
| 0.0 | 0.3 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 1.7 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.5 | GO:2000193 | positive regulation of fatty acid transport(GO:2000193) |
| 0.0 | 0.3 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.0 | 1.6 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.3 | GO:0021559 | trigeminal nerve development(GO:0021559) |
| 0.0 | 2.5 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.3 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.1 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.4 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.3 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.3 | GO:1902855 | nonmotile primary cilium assembly(GO:0035058) regulation of nonmotile primary cilium assembly(GO:1902855) positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.5 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.0 | GO:2000407 | regulation of T cell extravasation(GO:2000407) positive regulation of T cell extravasation(GO:2000409) |
| 0.0 | 0.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 1.2 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.9 | GO:0071482 | cellular response to light stimulus(GO:0071482) |
| 0.0 | 0.5 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 1.4 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.9 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 1.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 2.0 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 1.0 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.0 | 0.3 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 5.9 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 1.0 | 5.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.8 | 3.4 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.8 | 8.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.8 | 4.6 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.7 | 3.3 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.7 | 2.0 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.5 | 2.6 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.5 | 2.6 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.5 | 4.0 | GO:0042825 | TAP complex(GO:0042825) |
| 0.5 | 6.4 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.5 | 5.3 | GO:0070578 | micro-ribonucleoprotein complex(GO:0035068) RISC-loading complex(GO:0070578) |
| 0.5 | 2.4 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.4 | 1.3 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.4 | 8.9 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.4 | 3.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.4 | 3.4 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.4 | 4.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.4 | 10.7 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.4 | 1.1 | GO:0060473 | cortical granule(GO:0060473) |
| 0.3 | 1.4 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.3 | 0.9 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.3 | 1.9 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.2 | 2.0 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.2 | 3.4 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.2 | 2.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.2 | 1.5 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.2 | 2.8 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.2 | 3.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 1.9 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.2 | 2.3 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.2 | 1.5 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.2 | 1.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 1.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.2 | 1.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.2 | 2.0 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.2 | 1.7 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.2 | 8.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.2 | 1.8 | GO:0090571 | SMN-Sm protein complex(GO:0034719) RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.2 | 5.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 2.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.4 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 1.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 1.1 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 0.8 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.1 | 2.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 0.4 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.1 | 1.7 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 1.4 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 1.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 3.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 2.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 2.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 1.8 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.1 | 1.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 5.6 | GO:0000791 | euchromatin(GO:0000791) |
| 0.1 | 2.0 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 2.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 1.0 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.9 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 3.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 1.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 5.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 1.8 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 0.4 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.1 | 1.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.5 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 1.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.7 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.4 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 1.6 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 0.6 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 0.6 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 2.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 3.7 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 3.3 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 6.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 28.2 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.8 | GO:0005682 | U5 snRNP(GO:0005682) U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.6 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 1.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.6 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 4.5 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.9 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.0 | 0.2 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.8 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 2.5 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 4.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 2.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 5.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 2.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 2.2 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.9 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.4 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 14.1 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 1.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 32.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.8 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.6 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.6 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 1.7 | 8.3 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 1.3 | 4.0 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 1.0 | 6.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 1.0 | 6.7 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.9 | 2.6 | GO:0046911 | metal chelating activity(GO:0046911) |
| 0.8 | 5.6 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.8 | 3.2 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.8 | 4.6 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.8 | 2.3 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.7 | 2.1 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.6 | 1.9 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.6 | 3.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.6 | 5.9 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.6 | 2.3 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.5 | 2.2 | GO:0051435 | BH4 domain binding(GO:0051435) |
| 0.5 | 6.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.4 | 8.9 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.4 | 1.3 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.4 | 1.3 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.4 | 4.8 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.4 | 1.2 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.4 | 1.1 | GO:0036461 | BLOC-2 complex binding(GO:0036461) |
| 0.4 | 3.0 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.4 | 2.6 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.4 | 1.1 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.4 | 1.1 | GO:0070002 | glutamic-type peptidase activity(GO:0070002) |
| 0.4 | 2.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.3 | 1.7 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 0.3 | 1.4 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.3 | 1.4 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.3 | 1.0 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.3 | 1.6 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.3 | 1.5 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.3 | 1.5 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.3 | 0.8 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.3 | 2.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.3 | 2.4 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.3 | 1.3 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.2 | 1.7 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.2 | 1.0 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.2 | 3.4 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.2 | 5.5 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.2 | 2.5 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.2 | 2.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 1.5 | GO:0043426 | MRF binding(GO:0043426) |
| 0.2 | 1.7 | GO:0015099 | nickel cation transmembrane transporter activity(GO:0015099) |
| 0.2 | 2.3 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.2 | 0.6 | GO:0051747 | cytosine C-5 DNA demethylase activity(GO:0051747) |
| 0.2 | 1.2 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.2 | 4.7 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 7.6 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.2 | 0.9 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.2 | 8.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 3.0 | GO:0001727 | lipid kinase activity(GO:0001727) diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 7.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.2 | 8.4 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.2 | 2.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 1.8 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.2 | 0.5 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.2 | 1.9 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.2 | 3.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 2.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.2 | 1.5 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 2.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 1.0 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.1 | 1.4 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 0.7 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.1 | 3.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.8 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 7.4 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.1 | 1.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 1.8 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.1 | 1.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 1.7 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 1.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 1.0 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 4.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 1.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 1.5 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 0.7 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 2.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 2.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 2.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.6 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.1 | 1.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 5.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.4 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 3.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 5.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 1.1 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 5.9 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.1 | 0.4 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 1.7 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 0.3 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.1 | 0.4 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.1 | 0.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.9 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.6 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.1 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 5.7 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 0.4 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 3.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 2.1 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 11.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 3.9 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 2.8 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 0.1 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.1 | 2.4 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 1.2 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 1.4 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 6.0 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 2.0 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 2.2 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.6 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 6.8 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 2.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.6 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0018271 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.0 | 1.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.8 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 4.1 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 1.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.4 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.6 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 4.6 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 1.5 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.5 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 11.9 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.2 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 1.3 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.1 | GO:0044390 | ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 1.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 7.3 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.4 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 5.2 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 2.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.6 | GO:0042393 | histone binding(GO:0042393) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 23.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.3 | 4.4 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.3 | 4.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.3 | 18.0 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.2 | 19.7 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.2 | 1.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 3.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 9.6 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 3.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 2.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 2.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 6.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 2.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 1.0 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 3.8 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 8.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 7.1 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 2.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 0.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 2.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 2.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 2.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 2.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 2.2 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 11.9 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.5 | 8.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.4 | 8.0 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.4 | 7.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.4 | 4.6 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.3 | 2.4 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.3 | 12.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.3 | 6.8 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.3 | 8.0 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 4.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 6.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 4.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.2 | 6.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 2.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.2 | 5.9 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 5.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.8 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 1.4 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.1 | 2.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 2.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 3.3 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 2.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 6.2 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.1 | 2.0 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 10.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 4.7 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 7.2 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.1 | 1.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 4.0 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 2.7 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.1 | 2.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 0.4 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 2.0 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 4.7 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.1 | 2.5 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.4 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 2.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.3 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 3.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 2.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 3.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.6 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.2 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |