Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Rara
Z-value: 0.64
Transcription factors associated with Rara
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rara
|
ENSMUSG00000037992.17 | Rara |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rara | mm39_v1_chr11_+_98818640_98818650 | 0.29 | 1.3e-02 | Click! |
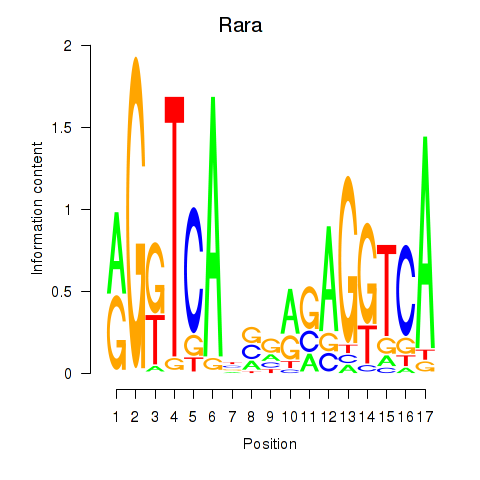
Activity profile of Rara motif
Sorted Z-values of Rara motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Rara
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_16716989 | 5.22 |
ENSMUST00000206129.3
|
Gm42372
|
predicted gene, 42372 |
| chr18_-_39652468 | 4.07 |
ENSMUST00000237944.2
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr9_-_83028523 | 3.50 |
ENSMUST00000187193.7
ENSMUST00000185315.7 |
Hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr9_-_83028654 | 3.39 |
ENSMUST00000161796.9
ENSMUST00000162246.9 |
Hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr1_-_172034354 | 2.88 |
ENSMUST00000013842.12
ENSMUST00000111247.8 |
Pea15a
|
phosphoprotein enriched in astrocytes 15A |
| chr1_-_172034251 | 2.75 |
ENSMUST00000155109.2
|
Pea15a
|
phosphoprotein enriched in astrocytes 15A |
| chr6_+_67873135 | 2.74 |
ENSMUST00000103310.3
|
Igkv14-126
|
immunoglobulin kappa variable 14-126 |
| chr7_+_4925781 | 2.70 |
ENSMUST00000207527.2
ENSMUST00000207687.2 ENSMUST00000208754.2 |
Nat14
|
N-acetyltransferase 14 |
| chr11_+_4936824 | 2.69 |
ENSMUST00000109897.8
ENSMUST00000009234.16 |
Ap1b1
|
adaptor protein complex AP-1, beta 1 subunit |
| chr16_-_19079594 | 2.68 |
ENSMUST00000103752.3
ENSMUST00000197518.2 |
Iglv2
|
immunoglobulin lambda variable 2 |
| chr8_+_69661683 | 2.68 |
ENSMUST00000212681.2
ENSMUST00000212312.2 |
Zfp930
|
zinc finger protein 930 |
| chr17_+_34812361 | 2.50 |
ENSMUST00000174532.2
|
Pbx2
|
pre B cell leukemia homeobox 2 |
| chr17_-_27816151 | 2.48 |
ENSMUST00000231742.2
|
Nudt3
|
nudix (nucleotide diphosphate linked moiety X)-type motif 3 |
| chrX_-_104919201 | 2.48 |
ENSMUST00000198209.2
|
Atrx
|
ATRX, chromatin remodeler |
| chr4_-_123458465 | 2.25 |
ENSMUST00000238731.2
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr7_-_141009264 | 2.22 |
ENSMUST00000164387.2
ENSMUST00000137488.2 ENSMUST00000084436.10 |
Cend1
|
cell cycle exit and neuronal differentiation 1 |
| chr17_-_56440817 | 2.21 |
ENSMUST00000167545.3
|
Sema6b
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
| chr6_+_41107047 | 2.20 |
ENSMUST00000103271.2
|
Trbv13-3
|
T cell receptor beta, variable 13-3 |
| chr7_-_141009346 | 2.18 |
ENSMUST00000124444.2
|
Cend1
|
cell cycle exit and neuronal differentiation 1 |
| chr6_+_68233361 | 2.16 |
ENSMUST00000103320.3
|
Igkv14-111
|
immunoglobulin kappa variable 14-111 |
| chr12_-_115157739 | 2.14 |
ENSMUST00000103525.3
|
Ighv1-54
|
immunoglobulin heavy variable V1-54 |
| chr12_-_115832846 | 2.10 |
ENSMUST00000199373.2
|
Ighv1-78
|
immunoglobulin heavy variable 1-78 |
| chr11_-_69728560 | 1.97 |
ENSMUST00000108634.9
|
Nlgn2
|
neuroligin 2 |
| chr6_+_30723540 | 1.95 |
ENSMUST00000141130.2
ENSMUST00000115127.8 |
Mest
|
mesoderm specific transcript |
| chr6_+_114108190 | 1.92 |
ENSMUST00000032451.9
|
Slc6a11
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 11 |
| chr12_-_108859123 | 1.86 |
ENSMUST00000161154.2
ENSMUST00000161410.8 |
Wars
|
tryptophanyl-tRNA synthetase |
| chr9_-_108903117 | 1.84 |
ENSMUST00000161521.8
ENSMUST00000045011.9 |
Atrip
|
ATR interacting protein |
| chr5_+_14075281 | 1.80 |
ENSMUST00000073957.8
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chrX_-_104918911 | 1.75 |
ENSMUST00000200471.2
|
Atrx
|
ATRX, chromatin remodeler |
| chr6_+_68495964 | 1.75 |
ENSMUST00000199510.5
ENSMUST00000103325.3 |
Igkv14-100
|
immunoglobulin kappa chain variable 14-100 |
| chr1_+_133965228 | 1.68 |
ENSMUST00000162779.2
|
Fmod
|
fibromodulin |
| chr8_+_106877025 | 1.66 |
ENSMUST00000212963.2
ENSMUST00000034377.8 |
Pla2g15
|
phospholipase A2, group XV |
| chr2_+_181007177 | 1.66 |
ENSMUST00000108807.9
|
Zgpat
|
zinc finger, CCCH-type with G patch domain |
| chr9_-_45896663 | 1.65 |
ENSMUST00000214179.2
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr5_+_135835713 | 1.59 |
ENSMUST00000126232.8
|
Srrm3
|
serine/arginine repetitive matrix 3 |
| chr4_+_32238712 | 1.58 |
ENSMUST00000108180.9
|
Bach2
|
BTB and CNC homology, basic leucine zipper transcription factor 2 |
| chr10_-_8638215 | 1.51 |
ENSMUST00000212553.2
|
Sash1
|
SAM and SH3 domain containing 1 |
| chr14_-_70043079 | 1.40 |
ENSMUST00000022665.4
|
Rhobtb2
|
Rho-related BTB domain containing 2 |
| chr12_-_114687825 | 1.36 |
ENSMUST00000194968.6
ENSMUST00000103506.3 |
Ighv1-20
|
immunoglobulin heavy variable V1-20 |
| chr2_+_26209755 | 1.33 |
ENSMUST00000066889.13
|
Gpsm1
|
G-protein signalling modulator 1 (AGS3-like, C. elegans) |
| chr12_-_114355789 | 1.32 |
ENSMUST00000103486.2
|
Ighv6-3
|
immunoglobulin heavy variable 6-3 |
| chr6_+_68026941 | 1.32 |
ENSMUST00000103316.2
|
Igkv9-120
|
immunoglobulin kappa chain variable 9-120 |
| chr3_-_36529753 | 1.29 |
ENSMUST00000199478.2
|
Anxa5
|
annexin A5 |
| chr6_-_13677928 | 1.25 |
ENSMUST00000203078.2
ENSMUST00000045235.8 |
Bmt2
|
base methyltransferase of 25S rRNA 2 |
| chr14_-_70864666 | 1.24 |
ENSMUST00000022694.17
|
Dmtn
|
dematin actin binding protein |
| chrX_+_73298342 | 1.23 |
ENSMUST00000096424.11
|
Emd
|
emerin |
| chr14_+_76725876 | 1.21 |
ENSMUST00000101618.9
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr3_+_151916106 | 1.13 |
ENSMUST00000199202.5
ENSMUST00000200524.5 ENSMUST00000198227.5 ENSMUST00000196739.5 ENSMUST00000196695.5 ENSMUST00000106121.6 |
Fubp1
|
far upstream element (FUSE) binding protein 1 |
| chr13_+_83720457 | 1.07 |
ENSMUST00000196730.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chrX_-_74918122 | 1.07 |
ENSMUST00000033547.14
|
Pls3
|
plastin 3 (T-isoform) |
| chr13_+_83720484 | 1.05 |
ENSMUST00000196207.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr1_-_14374794 | 1.03 |
ENSMUST00000190337.7
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr4_+_48045143 | 1.02 |
ENSMUST00000030025.10
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr4_-_128503774 | 0.99 |
ENSMUST00000141040.2
ENSMUST00000147876.2 ENSMUST00000097877.9 |
Zscan20
|
zinc finger and SCAN domains 20 |
| chr6_-_54949587 | 0.99 |
ENSMUST00000060655.15
|
Nod1
|
nucleotide-binding oligomerization domain containing 1 |
| chr4_-_46991842 | 0.92 |
ENSMUST00000107749.4
|
Gabbr2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr8_+_91681550 | 0.92 |
ENSMUST00000210947.2
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr14_-_75185281 | 0.87 |
ENSMUST00000088970.7
ENSMUST00000228252.2 |
Lrch1
|
leucine-rich repeats and calponin homology (CH) domain containing 1 |
| chr9_-_45896110 | 0.86 |
ENSMUST00000215060.2
ENSMUST00000213853.2 ENSMUST00000216334.2 |
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr7_-_16790594 | 0.85 |
ENSMUST00000037762.11
|
Hif3a
|
hypoxia inducible factor 3, alpha subunit |
| chr9_-_44624496 | 0.83 |
ENSMUST00000144251.8
ENSMUST00000156918.8 |
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr11_-_97591150 | 0.80 |
ENSMUST00000018681.14
|
Pcgf2
|
polycomb group ring finger 2 |
| chr2_+_169475436 | 0.79 |
ENSMUST00000109157.2
|
Tshz2
|
teashirt zinc finger family member 2 |
| chr1_-_14374842 | 0.79 |
ENSMUST00000188857.7
ENSMUST00000185453.7 |
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr4_-_152402915 | 0.77 |
ENSMUST00000170820.3
ENSMUST00000076183.12 |
Rnf207
|
ring finger protein 207 |
| chr2_+_120807498 | 0.73 |
ENSMUST00000067582.14
|
Tmem62
|
transmembrane protein 62 |
| chr9_-_56325344 | 0.62 |
ENSMUST00000061552.15
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr6_+_18848600 | 0.61 |
ENSMUST00000201141.3
|
Lsm8
|
LSM8 homolog, U6 small nuclear RNA associated |
| chr7_-_30156826 | 0.60 |
ENSMUST00000045817.14
|
Kirrel2
|
kirre like nephrin family adhesion molecule 2 |
| chr3_+_79791798 | 0.58 |
ENSMUST00000118853.8
ENSMUST00000145992.2 |
Gask1b
|
golgi associated kinase 1B |
| chr4_+_150938376 | 0.57 |
ENSMUST00000073600.9
|
Errfi1
|
ERBB receptor feedback inhibitor 1 |
| chr15_-_103231921 | 0.56 |
ENSMUST00000229551.2
|
Zfp385a
|
zinc finger protein 385A |
| chr9_-_45896075 | 0.56 |
ENSMUST00000217636.2
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr5_-_105387395 | 0.51 |
ENSMUST00000065588.7
|
Gbp10
|
guanylate-binding protein 10 |
| chr6_+_18848570 | 0.51 |
ENSMUST00000056398.11
|
Lsm8
|
LSM8 homolog, U6 small nuclear RNA associated |
| chr14_+_58308004 | 0.49 |
ENSMUST00000165526.9
|
Fgf9
|
fibroblast growth factor 9 |
| chr1_-_163231196 | 0.48 |
ENSMUST00000045138.6
|
Gorab
|
golgin, RAB6-interacting |
| chr15_-_101694895 | 0.43 |
ENSMUST00000071104.6
|
Krt72
|
keratin 72 |
| chr14_+_20724378 | 0.38 |
ENSMUST00000224492.2
ENSMUST00000223751.2 ENSMUST00000225108.2 ENSMUST00000224754.2 |
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae) |
| chr3_-_103716593 | 0.38 |
ENSMUST00000063502.13
ENSMUST00000106832.2 ENSMUST00000106834.8 ENSMUST00000029435.15 |
Dclre1b
|
DNA cross-link repair 1B |
| chr14_+_20724366 | 0.36 |
ENSMUST00000048657.10
|
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae) |
| chr2_+_5850053 | 0.36 |
ENSMUST00000127116.7
ENSMUST00000194933.2 |
Nudt5
|
nudix (nucleoside diphosphate linked moiety X)-type motif 5 |
| chr6_+_115578863 | 0.35 |
ENSMUST00000000449.9
|
Mkrn2
|
makorin, ring finger protein, 2 |
| chr3_+_79792238 | 0.27 |
ENSMUST00000135021.2
|
Gask1b
|
golgi associated kinase 1B |
| chr2_-_181007099 | 0.27 |
ENSMUST00000108808.8
ENSMUST00000170190.8 ENSMUST00000127988.8 |
Arfrp1
|
ADP-ribosylation factor related protein 1 |
| chr8_-_120505104 | 0.27 |
ENSMUST00000212534.2
ENSMUST00000049156.7 |
Meak7
|
MTOR associated protein , eak-7 homolog |
| chr6_-_39787813 | 0.26 |
ENSMUST00000114797.2
ENSMUST00000031978.9 |
Mrps33
|
mitochondrial ribosomal protein S33 |
| chr9_-_96513529 | 0.25 |
ENSMUST00000034984.8
|
Rasa2
|
RAS p21 protein activator 2 |
| chr17_+_34131696 | 0.25 |
ENSMUST00000174146.3
|
Daxx
|
Fas death domain-associated protein |
| chr1_+_157286124 | 0.25 |
ENSMUST00000193791.6
ENSMUST00000046743.11 ENSMUST00000119891.7 |
Cryzl2
|
crystallin zeta like 2 |
| chr2_+_131104389 | 0.22 |
ENSMUST00000150843.9
|
Pank2
|
pantothenate kinase 2 |
| chr7_+_15484302 | 0.20 |
ENSMUST00000098802.10
ENSMUST00000173053.2 |
Obox5
|
oocyte specific homeobox 5 |
| chr7_-_18390645 | 0.20 |
ENSMUST00000094793.12
ENSMUST00000182128.2 |
Psg21
|
pregnancy-specific glycoprotein 21 |
| chr7_+_17447163 | 0.18 |
ENSMUST00000081907.8
|
Ceacam5
|
carcinoembryonic antigen-related cell adhesion molecule 5 |
| chr4_+_32238950 | 0.14 |
ENSMUST00000037416.13
|
Bach2
|
BTB and CNC homology, basic leucine zipper transcription factor 2 |
| chr3_-_87965757 | 0.12 |
ENSMUST00000029708.8
|
Naxe
|
NAD(P)HX epimerase |
| chr8_+_11606056 | 0.11 |
ENSMUST00000210740.2
ENSMUST00000210670.2 ENSMUST00000054399.6 ENSMUST00000209646.2 |
Ing1
|
inhibitor of growth family, member 1 |
| chr1_+_93096316 | 0.08 |
ENSMUST00000138595.3
|
Crocc2
|
ciliary rootlet coiled-coil, rootletin family member 2 |
| chr7_+_15484056 | 0.05 |
ENSMUST00000173455.8
|
Obox5
|
oocyte specific homeobox 5 |
| chr5_+_53424471 | 0.05 |
ENSMUST00000147148.5
|
Smim20
|
small integral membrane protein 20 |
| chr14_-_30075424 | 0.04 |
ENSMUST00000224198.3
ENSMUST00000238675.2 ENSMUST00000112249.10 ENSMUST00000224785.3 |
Cacna1d
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr7_+_15122873 | 0.04 |
ENSMUST00000181001.8
ENSMUST00000174076.8 ENSMUST00000174305.8 |
Obox1
Obox2
|
oocyte specific homeobox 1 oocyte specific homeobox 2 |
| chr11_-_5211558 | 0.04 |
ENSMUST00000020662.15
|
Kremen1
|
kringle containing transmembrane protein 1 |
| chr11_+_3438274 | 0.03 |
ENSMUST00000064265.13
|
Pla2g3
|
phospholipase A2, group III |
| chr5_+_53747556 | 0.02 |
ENSMUST00000037618.13
ENSMUST00000201912.4 |
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.4 | GO:0021941 | radial glia guided migration of cerebellar granule cell(GO:0021933) negative regulation of cerebellar granule cell precursor proliferation(GO:0021941) |
| 0.7 | 4.2 | GO:0035128 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.7 | 2.0 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.6 | 2.5 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.6 | 4.1 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.6 | 1.7 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.5 | 1.9 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.3 | 1.5 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.3 | 0.8 | GO:1903116 | positive regulation of actin filament-based movement(GO:1903116) |
| 0.3 | 1.0 | GO:1900623 | positive regulation of mast cell cytokine production(GO:0032765) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.2 | 1.3 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.2 | 1.2 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.2 | 2.1 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.2 | 5.6 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.2 | 1.8 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.6 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 1.0 | GO:0002604 | regulation of dendritic cell antigen processing and presentation(GO:0002604) positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.1 | 2.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 0.6 | GO:1903243 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.1 | 0.4 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 2.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 1.7 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 6.9 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 1.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.8 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.2 | GO:0072737 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.0 | 6.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 2.5 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 1.8 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) |
| 0.0 | 3.1 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 0.7 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 1.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.5 | GO:1901620 | regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901620) |
| 0.0 | 1.9 | GO:0010883 | regulation of lipid storage(GO:0010883) |
| 0.0 | 0.5 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 1.1 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.1 | GO:0006014 | D-ribose metabolic process(GO:0006014) pentose catabolic process(GO:0019323) |
| 0.0 | 0.2 | GO:1904251 | regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.9 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 3.1 | GO:0002377 | immunoglobulin production(GO:0002377) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.2 | GO:1990707 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.2 | 0.9 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.2 | 1.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 1.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 2.0 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 1.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 0.8 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 6.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 2.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.6 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 5.6 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 1.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 3.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 6.3 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 1.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.4 | GO:0045095 | keratin filament(GO:0045095) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.1 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.6 | 4.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.5 | 1.9 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.4 | 3.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.4 | 2.5 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.3 | 1.9 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.2 | 1.3 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.2 | 4.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 1.7 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.2 | 6.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 0.9 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 2.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 1.0 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 2.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.8 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 5.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 6.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 1.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.4 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.0 | 1.5 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 1.0 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 2.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 1.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 2.7 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 1.8 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 1.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.0 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.0 | 0.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.8 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 1.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 3.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 2.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 2.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.8 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 4.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 1.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.9 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 2.1 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.8 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.7 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 1.0 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.5 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |