Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Rarb
Z-value: 0.91
Transcription factors associated with Rarb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rarb
|
ENSMUSG00000017491.10 | Rarb |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rarb | mm39_v1_chr14_+_5894220_5894254 | 0.09 | 4.5e-01 | Click! |
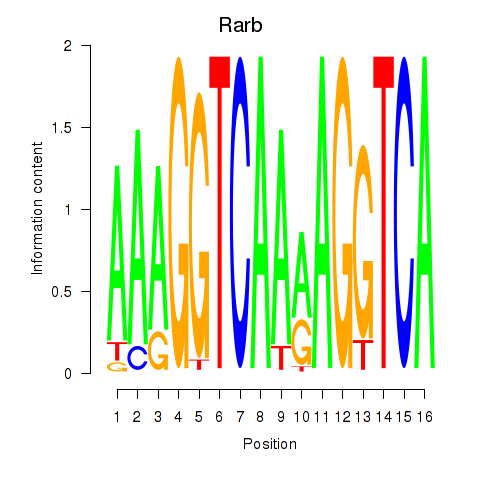
Activity profile of Rarb motif
Sorted Z-values of Rarb motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Rarb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_8027640 | 15.16 |
ENSMUST00000171271.8
ENSMUST00000037811.13 |
Apob
|
apolipoprotein B |
| chr12_+_8027767 | 15.02 |
ENSMUST00000037520.14
|
Apob
|
apolipoprotein B |
| chr5_-_87054796 | 14.73 |
ENSMUST00000031181.16
ENSMUST00000113333.2 |
Ugt2b34
|
UDP glucuronosyltransferase 2 family, polypeptide B34 |
| chr5_-_87288177 | 12.22 |
ENSMUST00000067790.7
|
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr6_-_85797946 | 9.79 |
ENSMUST00000032074.5
|
Nat8f5
|
N-acetyltransferase 8 (GCN5-related) family member 5 |
| chr7_+_35148461 | 8.59 |
ENSMUST00000118969.8
|
Slc7a9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 |
| chr19_+_44980565 | 8.30 |
ENSMUST00000179305.2
|
Sema4g
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr7_+_35148579 | 8.29 |
ENSMUST00000032703.10
|
Slc7a9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 |
| chr7_+_35148188 | 8.27 |
ENSMUST00000118383.8
|
Slc7a9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 |
| chr11_+_76795346 | 7.36 |
ENSMUST00000072633.4
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr11_+_76795292 | 6.58 |
ENSMUST00000142166.8
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr6_-_85797917 | 5.72 |
ENSMUST00000174143.2
|
Nat8f6
|
N-acetyltransferase 8 (GCN5-related) family member 6 |
| chr17_+_12803019 | 5.06 |
ENSMUST00000046959.9
ENSMUST00000233066.2 |
Slc22a2
|
solute carrier family 22 (organic cation transporter), member 2 |
| chr5_-_87074380 | 4.10 |
ENSMUST00000031183.3
|
Ugt2b1
|
UDP glucuronosyltransferase 2 family, polypeptide B1 |
| chr1_+_4878046 | 3.01 |
ENSMUST00000027036.11
ENSMUST00000150971.8 ENSMUST00000119612.9 ENSMUST00000137887.8 ENSMUST00000115529.8 |
Lypla1
|
lysophospholipase 1 |
| chr7_-_115630282 | 2.68 |
ENSMUST00000206034.2
ENSMUST00000106612.8 |
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr5_+_93045837 | 2.31 |
ENSMUST00000113051.9
|
Shroom3
|
shroom family member 3 |
| chr13_-_25454058 | 2.27 |
ENSMUST00000057866.13
|
Nrsn1
|
neurensin 1 |
| chr11_-_71092282 | 1.47 |
ENSMUST00000108515.9
|
Nlrp1b
|
NLR family, pyrin domain containing 1B |
| chr2_+_3771709 | 1.14 |
ENSMUST00000177037.2
|
Fam107b
|
family with sequence similarity 107, member B |
| chr11_-_71092124 | 1.11 |
ENSMUST00000108514.10
|
Nlrp1b
|
NLR family, pyrin domain containing 1B |
| chr8_+_36956345 | 1.04 |
ENSMUST00000171777.2
|
Trmt9b
|
tRNA methyltransferase 9B |
| chr3_+_108093645 | 0.57 |
ENSMUST00000050909.7
ENSMUST00000106659.3 ENSMUST00000106656.2 |
Amigo1
|
adhesion molecule with Ig like domain 1 |
| chr1_+_24216691 | 0.54 |
ENSMUST00000054588.15
|
Col9a1
|
collagen, type IX, alpha 1 |
| chr10_-_81332163 | 0.54 |
ENSMUST00000020463.14
|
Ncln
|
nicalin |
| chr7_+_45267024 | 0.30 |
ENSMUST00000008605.6
ENSMUST00000209379.2 |
Fut1
|
fucosyltransferase 1 |
| chr14_+_51648277 | 0.22 |
ENSMUST00000159674.4
ENSMUST00000163019.9 ENSMUST00000233394.2 ENSMUST00000232951.2 ENSMUST00000232797.2 ENSMUST00000233766.2 ENSMUST00000233848.2 ENSMUST00000233280.2 ENSMUST00000233920.2 ENSMUST00000233126.2 ENSMUST00000233651.2 |
Vmn2r88
|
vomeronasal 2, receptor 88 |
| chr16_+_38167352 | 0.18 |
ENSMUST00000050273.9
ENSMUST00000120495.2 ENSMUST00000119704.2 |
Cox17
Gm21987
|
cytochrome c oxidase assembly protein 17, copper chaperone predicted gene 21987 |
| chr3_-_52012462 | 0.06 |
ENSMUST00000121440.4
|
Maml3
|
mastermind like transcriptional coactivator 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 25.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 2.2 | 30.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 1.4 | 4.1 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.6 | 2.6 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 0.5 | 3.0 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.3 | 5.1 | GO:0051608 | quaternary ammonium group transport(GO:0015697) histamine transport(GO:0051608) |
| 0.2 | 2.7 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.2 | 9.8 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.1 | 8.3 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 0.2 | GO:1904959 | regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.0 | 0.6 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.5 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 2.3 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 | 0.3 | GO:0036065 | fucosylation(GO:0036065) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.1 | 30.2 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.4 | 2.6 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.2 | 25.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.5 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 12.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 5.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 25.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 2.3 | 30.2 | GO:0035473 | lipase binding(GO:0035473) |
| 1.7 | 5.1 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.8 | 31.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.3 | 3.0 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.3 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.1 | 6.9 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 30.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 8.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 30.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.5 | 25.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.4 | 5.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 1.3 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |