Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
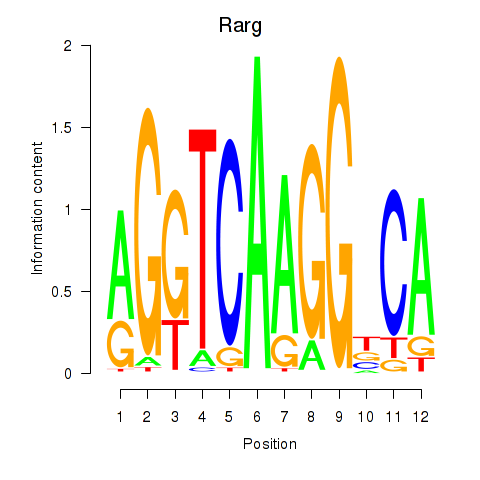
Results for Rarg
Z-value: 1.82
Transcription factors associated with Rarg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rarg
|
ENSMUSG00000001288.16 | Rarg |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rarg | mm39_v1_chr15_-_102165740_102165806 | -0.53 | 1.5e-06 | Click! |
Activity profile of Rarg motif
Sorted Z-values of Rarg motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Rarg
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_4815645 | 11.98 |
ENSMUST00000229321.2
ENSMUST00000230362.2 ENSMUST00000170323.3 |
Septin12
|
septin 12 |
| chr11_-_69712970 | 7.88 |
ENSMUST00000045771.7
|
Spem1
|
sperm maturation 1 |
| chr14_-_75830550 | 7.41 |
ENSMUST00000164082.9
ENSMUST00000169658.9 |
Cby2
|
chibby family member 2 |
| chr12_-_115944754 | 7.09 |
ENSMUST00000103551.2
|
Ighv1-84
|
immunoglobulin heavy variable 1-84 |
| chr7_+_46510677 | 7.09 |
ENSMUST00000211784.2
ENSMUST00000210585.2 ENSMUST00000148565.8 |
Ldhc
|
lactate dehydrogenase C |
| chr12_-_113958518 | 7.04 |
ENSMUST00000103467.2
|
Ighv14-2
|
immunoglobulin heavy variable 14-2 |
| chr7_+_46510627 | 6.68 |
ENSMUST00000014545.11
|
Ldhc
|
lactate dehydrogenase C |
| chr7_+_46510831 | 6.52 |
ENSMUST00000126004.3
|
Ldhc
|
lactate dehydrogenase C |
| chrX_-_100307592 | 6.41 |
ENSMUST00000101358.3
|
Gm614
|
predicted gene 614 |
| chr15_+_76178542 | 6.37 |
ENSMUST00000163991.4
|
Smpd5
|
sphingomyelin phosphodiesterase 5 |
| chr4_-_154752320 | 6.23 |
ENSMUST00000060062.5
|
Actrt2
|
actin-related protein T2 |
| chr8_-_106706035 | 6.21 |
ENSMUST00000034371.9
|
Dpep3
|
dipeptidase 3 |
| chr9_+_44893077 | 6.17 |
ENSMUST00000034602.9
|
Cd3d
|
CD3 antigen, delta polypeptide |
| chr7_+_43910845 | 6.15 |
ENSMUST00000124863.4
|
Gm15517
|
predicted gene 15517 |
| chr12_-_114547622 | 6.14 |
ENSMUST00000193893.6
ENSMUST00000103498.3 |
Ighv1-9
|
immunoglobulin heavy variable V1-9 |
| chr12_+_103281188 | 6.14 |
ENSMUST00000189885.2
ENSMUST00000179363.2 |
Fam181a
|
family with sequence similarity 181, member A |
| chrX_-_7834057 | 6.12 |
ENSMUST00000033502.14
|
Gata1
|
GATA binding protein 1 |
| chr3_-_83947416 | 5.93 |
ENSMUST00000192095.6
ENSMUST00000191758.6 ENSMUST00000052342.9 |
Tmem131l
|
transmembrane 131 like |
| chr16_+_90535212 | 5.87 |
ENSMUST00000038197.3
|
Mrap
|
melanocortin 2 receptor accessory protein |
| chr16_+_9988080 | 5.49 |
ENSMUST00000121292.8
ENSMUST00000044103.6 |
Rpl39l
|
ribosomal protein L39-like |
| chr12_-_115083839 | 5.48 |
ENSMUST00000103521.3
|
Ighv1-50
|
immunoglobulin heavy variable 1-50 |
| chr10_-_75658355 | 5.44 |
ENSMUST00000160211.2
|
Gstt4
|
glutathione S-transferase, theta 4 |
| chr3_+_96432479 | 5.42 |
ENSMUST00000049208.11
|
Hjv
|
hemojuvelin BMP co-receptor |
| chr13_-_24945423 | 5.31 |
ENSMUST00000176890.8
ENSMUST00000175689.8 |
Gmnn
|
geminin |
| chr12_-_115172211 | 5.28 |
ENSMUST00000103526.3
|
Ighv1-55
|
immunoglobulin heavy variable 1-55 |
| chrY_+_818646 | 5.25 |
ENSMUST00000190013.7
ENSMUST00000115894.3 |
Uba1y
|
ubiquitin-activating enzyme, Chr Y |
| chr5_+_137759934 | 5.24 |
ENSMUST00000110983.3
ENSMUST00000031738.5 |
Tsc22d4
|
TSC22 domain family, member 4 |
| chr12_-_115122455 | 5.20 |
ENSMUST00000103523.2
|
Ighv1-53
|
immunoglobulin heavy variable 1-53 |
| chr7_+_24596806 | 5.17 |
ENSMUST00000003469.8
|
Cd79a
|
CD79A antigen (immunoglobulin-associated alpha) |
| chr16_-_10609959 | 5.11 |
ENSMUST00000037996.7
|
Prm2
|
protamine 2 |
| chr11_-_5753693 | 5.02 |
ENSMUST00000020768.4
|
Pgam2
|
phosphoglycerate mutase 2 |
| chr1_-_164988342 | 5.02 |
ENSMUST00000027859.12
|
Tbx19
|
T-box 19 |
| chrX_-_100312629 | 4.99 |
ENSMUST00000117736.2
|
Gm20489
|
predicted gene 20489 |
| chr12_-_115916604 | 4.98 |
ENSMUST00000196991.2
|
Ighv1-82
|
immunoglobulin heavy variable 1-82 |
| chr1_+_52047368 | 4.88 |
ENSMUST00000027277.7
|
Stat4
|
signal transducer and activator of transcription 4 |
| chr19_-_3979723 | 4.84 |
ENSMUST00000051803.8
|
Aldh3b1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr12_-_115299134 | 4.83 |
ENSMUST00000195359.6
ENSMUST00000103530.3 |
Ighv1-59
|
immunoglobulin heavy variable V1-59 |
| chr2_+_172863688 | 4.76 |
ENSMUST00000029014.16
|
Rbm38
|
RNA binding motif protein 38 |
| chr2_-_25507680 | 4.74 |
ENSMUST00000028309.4
|
Ccdc183
|
coiled-coil domain containing 183 |
| chr7_-_127805518 | 4.68 |
ENSMUST00000033049.9
|
Cox6a2
|
cytochrome c oxidase subunit 6A2 |
| chr11_+_117673107 | 4.66 |
ENSMUST00000050874.14
ENSMUST00000106334.9 |
Tmc8
|
transmembrane channel-like gene family 8 |
| chr7_+_43090206 | 4.64 |
ENSMUST00000040227.3
|
Cldnd2
|
claudin domain containing 2 |
| chr12_+_78240999 | 4.60 |
ENSMUST00000211288.3
|
Gm6657
|
predicted gene 6657 |
| chr8_+_72050292 | 4.60 |
ENSMUST00000143662.8
|
Niban3
|
niban apoptosis regulator 3 |
| chr1_+_160806194 | 4.50 |
ENSMUST00000064725.11
ENSMUST00000191936.2 |
Serpinc1
|
serine (or cysteine) peptidase inhibitor, clade C (antithrombin), member 1 |
| chr9_-_58277734 | 4.47 |
ENSMUST00000040217.6
|
Tbc1d21
|
TBC1 domain family, member 21 |
| chr17_-_32639936 | 4.45 |
ENSMUST00000170392.9
ENSMUST00000237165.2 ENSMUST00000235892.2 ENSMUST00000114455.3 |
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr17_-_28298721 | 4.42 |
ENSMUST00000233898.2
ENSMUST00000232798.2 |
Tcp11
|
t-complex protein 11 |
| chr12_-_114140482 | 4.37 |
ENSMUST00000103475.2
ENSMUST00000195706.2 |
Ighv14-4
|
immunoglobulin heavy variable 14-4 |
| chr10_-_80165257 | 4.36 |
ENSMUST00000020340.15
|
Pcsk4
|
proprotein convertase subtilisin/kexin type 4 |
| chr1_+_177635997 | 4.28 |
ENSMUST00000194528.3
|
Catspere1
|
cation channel sperm associated auxiliary subunit epsilon 1 |
| chr8_+_11890474 | 4.25 |
ENSMUST00000033909.14
ENSMUST00000209692.2 |
Tex29
|
testis expressed 29 |
| chr6_+_41661356 | 4.22 |
ENSMUST00000031900.6
|
Llcfc1
|
LLLL and CFNLAS motif containing 1 |
| chr1_+_153300874 | 4.22 |
ENSMUST00000042373.12
|
Shcbp1l
|
Shc SH2-domain binding protein 1-like |
| chr12_-_115587215 | 4.21 |
ENSMUST00000199933.5
ENSMUST00000103539.3 |
Ighv1-69
|
immunoglobulin heavy variable 1-69 |
| chr2_-_162929732 | 4.15 |
ENSMUST00000094653.6
|
Gtsf1l
|
gametocyte specific factor 1-like |
| chr7_+_26728499 | 4.15 |
ENSMUST00000075552.7
|
Cyp2a12
|
cytochrome P450, family 2, subfamily a, polypeptide 12 |
| chr17_-_35077089 | 4.13 |
ENSMUST00000153400.8
|
Cfb
|
complement factor B |
| chr3_+_98067940 | 4.12 |
ENSMUST00000198363.2
ENSMUST00000050342.5 |
Adam30
|
a disintegrin and metallopeptidase domain 30 |
| chr12_-_114621406 | 4.08 |
ENSMUST00000192077.2
|
Ighv1-15
|
immunoglobulin heavy variable 1-15 |
| chr7_+_44485704 | 4.07 |
ENSMUST00000033015.8
|
Il4i1
|
interleukin 4 induced 1 |
| chr1_-_156248716 | 4.06 |
ENSMUST00000178036.8
|
Axdnd1
|
axonemal dynein light chain domain containing 1 |
| chr7_+_43701714 | 4.03 |
ENSMUST00000079859.7
|
Klk1b27
|
kallikrein 1-related peptidase b27 |
| chr19_+_30210320 | 4.00 |
ENSMUST00000025797.7
|
Mbl2
|
mannose-binding lectin (protein C) 2 |
| chr15_+_76152276 | 3.94 |
ENSMUST00000074173.4
|
Spatc1
|
spermatogenesis and centriole associated 1 |
| chr6_+_121187636 | 3.92 |
ENSMUST00000032233.9
|
Tuba8
|
tubulin, alpha 8 |
| chr7_-_79443536 | 3.92 |
ENSMUST00000032760.6
|
Mesp1
|
mesoderm posterior 1 |
| chrX_+_106299484 | 3.86 |
ENSMUST00000101294.9
ENSMUST00000118820.8 ENSMUST00000120971.8 |
Gpr174
|
G protein-coupled receptor 174 |
| chr3_-_95310106 | 3.83 |
ENSMUST00000196239.2
|
4930558C23Rik
|
RIKEN cDNA 4930558C23 gene |
| chr9_-_106448182 | 3.76 |
ENSMUST00000085111.5
|
Iqcf4
|
IQ motif containing F4 |
| chr6_+_128864549 | 3.74 |
ENSMUST00000032519.12
ENSMUST00000159866.8 ENSMUST00000162666.4 ENSMUST00000160867.3 |
Clec2i
|
C-type lectin domain family 2, member i |
| chr7_+_28136861 | 3.73 |
ENSMUST00000108292.9
ENSMUST00000108289.8 |
Gmfg
|
glia maturation factor, gamma |
| chr6_+_126830102 | 3.71 |
ENSMUST00000202878.4
ENSMUST00000202574.2 |
Akap3
|
A kinase (PRKA) anchor protein 3 |
| chr12_-_84923252 | 3.70 |
ENSMUST00000163189.8
ENSMUST00000110254.9 ENSMUST00000002073.13 |
Ltbp2
|
latent transforming growth factor beta binding protein 2 |
| chr12_-_55033130 | 3.70 |
ENSMUST00000173433.8
ENSMUST00000173803.2 |
Baz1a
Gm20403
|
bromodomain adjacent to zinc finger domain 1A predicted gene 20403 |
| chr7_-_29426425 | 3.69 |
ENSMUST00000061193.4
|
Catsperg2
|
cation channel sperm associated auxiliary subunit gamma 2 |
| chr9_-_106421834 | 3.68 |
ENSMUST00010181660.1
ENSMUST00000215525.2 |
ENSMUSG00001074846.1
ENSMUSG00000118396.3
|
IQ motif containing F3 IQ motif containing F3 |
| chr7_+_30681287 | 3.66 |
ENSMUST00000128384.3
|
Fam187b
|
family with sequence similarity 187, member B |
| chr7_-_140859034 | 3.65 |
ENSMUST00000211667.2
ENSMUST00000167790.3 ENSMUST00000046156.13 |
Sct
|
secretin |
| chr6_-_48747767 | 3.62 |
ENSMUST00000204036.2
|
Gimap3
|
GTPase, IMAP family member 3 |
| chr15_-_71906051 | 3.61 |
ENSMUST00000159993.8
|
Col22a1
|
collagen, type XXII, alpha 1 |
| chr10_-_21943978 | 3.61 |
ENSMUST00000092672.6
|
4930444G20Rik
|
RIKEN cDNA 4930444G20 gene |
| chr8_-_13888389 | 3.58 |
ENSMUST00000071308.6
|
AF366264
|
cDNA sequence AF366264 |
| chr1_+_171745075 | 3.58 |
ENSMUST00000195656.6
ENSMUST00000171330.7 |
Slamf6
|
SLAM family member 6 |
| chr7_+_90075762 | 3.56 |
ENSMUST00000061391.9
|
Ccdc89
|
coiled-coil domain containing 89 |
| chr12_-_114752425 | 3.53 |
ENSMUST00000103510.2
|
Ighv1-26
|
immunoglobulin heavy variable 1-26 |
| chr12_-_115884332 | 3.53 |
ENSMUST00000103548.3
|
Ighv1-81
|
immunoglobulin heavy variable 1-81 |
| chr18_-_35795233 | 3.52 |
ENSMUST00000025209.12
ENSMUST00000096573.4 |
Spata24
|
spermatogenesis associated 24 |
| chr2_+_84810802 | 3.52 |
ENSMUST00000028467.6
|
Prg2
|
proteoglycan 2, bone marrow |
| chr19_-_4241034 | 3.50 |
ENSMUST00000237495.2
|
Tbc1d10c
|
TBC1 domain family, member 10c |
| chr1_-_192883642 | 3.49 |
ENSMUST00000192020.6
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr7_+_119927885 | 3.48 |
ENSMUST00000207220.2
ENSMUST00000121265.7 ENSMUST00000076272.5 |
Abca15
|
ATP-binding cassette, sub-family A (ABC1), member 15 |
| chr6_+_126830050 | 3.47 |
ENSMUST00000095440.9
|
Akap3
|
A kinase (PRKA) anchor protein 3 |
| chr2_+_148623929 | 3.46 |
ENSMUST00000028933.3
|
Cstdc1
|
cystatin domain containing 1 |
| chr5_+_136722995 | 3.46 |
ENSMUST00000197186.2
|
Myl10
|
myosin, light chain 10, regulatory |
| chr11_-_5865124 | 3.46 |
ENSMUST00000109823.9
ENSMUST00000109822.8 |
Gck
|
glucokinase |
| chr18_-_35795175 | 3.45 |
ENSMUST00000236574.2
ENSMUST00000236971.2 |
Spata24
|
spermatogenesis associated 24 |
| chr7_-_12731594 | 3.42 |
ENSMUST00000133977.3
|
Slc27a5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr19_-_4240984 | 3.39 |
ENSMUST00000045864.4
|
Tbc1d10c
|
TBC1 domain family, member 10c |
| chr2_+_172314433 | 3.38 |
ENSMUST00000029007.3
|
Fam209
|
family with sequence similarity 209 |
| chr16_+_17712061 | 3.36 |
ENSMUST00000046937.4
|
Tssk1
|
testis-specific serine kinase 1 |
| chr7_+_43625387 | 3.35 |
ENSMUST00000081399.4
|
Klk1b9
|
kallikrein 1-related peptidase b9 |
| chr3_-_88317601 | 3.34 |
ENSMUST00000193338.6
ENSMUST00000056370.13 |
Pmf1
|
polyamine-modulated factor 1 |
| chr4_-_6275629 | 3.34 |
ENSMUST00000029905.2
|
Cyp7a1
|
cytochrome P450, family 7, subfamily a, polypeptide 1 |
| chr17_+_57077258 | 3.30 |
ENSMUST00000168666.3
|
Prr22
|
proline rich 22 |
| chr8_+_71156071 | 3.26 |
ENSMUST00000212436.2
|
Iqcn
|
IQ motif containing N |
| chr9_+_110248815 | 3.25 |
ENSMUST00000035061.9
|
Ngp
|
neutrophilic granule protein |
| chr12_-_115722081 | 3.25 |
ENSMUST00000103541.3
|
Ighv1-72
|
immunoglobulin heavy variable 1-72 |
| chr2_+_152446244 | 3.23 |
ENSMUST00000058086.6
|
Defb36
|
defensin beta 36 |
| chr7_-_44828962 | 3.21 |
ENSMUST00000211004.2
ENSMUST00000179443.3 |
Gfy
|
golgi-associated olfactory signaling regulator |
| chr3_+_96736774 | 3.21 |
ENSMUST00000138014.8
|
Pdzk1
|
PDZ domain containing 1 |
| chr3_+_34074048 | 3.21 |
ENSMUST00000001620.13
|
Fxr1
|
fragile X mental retardation gene 1, autosomal homolog |
| chr12_-_114672701 | 3.20 |
ENSMUST00000103505.3
ENSMUST00000193855.2 |
Ighv1-19
|
immunoglobulin heavy variable V1-19 |
| chr12_-_115157739 | 3.20 |
ENSMUST00000103525.3
|
Ighv1-54
|
immunoglobulin heavy variable V1-54 |
| chr3_+_96736600 | 3.19 |
ENSMUST00000135031.8
|
Pdzk1
|
PDZ domain containing 1 |
| chr11_-_120441952 | 3.19 |
ENSMUST00000026121.3
|
Ppp1r27
|
protein phosphatase 1, regulatory subunit 27 |
| chr1_-_9770434 | 3.17 |
ENSMUST00000088658.11
|
Mybl1
|
myeloblastosis oncogene-like 1 |
| chr19_+_39275518 | 3.13 |
ENSMUST00000003137.15
|
Cyp2c29
|
cytochrome P450, family 2, subfamily c, polypeptide 29 |
| chr7_-_44646960 | 3.13 |
ENSMUST00000207443.2
ENSMUST00000207755.2 ENSMUST00000003290.12 |
Bcl2l12
|
BCL2-like 12 (proline rich) |
| chr11_+_32233511 | 3.11 |
ENSMUST00000093209.4
|
Hba-a1
|
hemoglobin alpha, adult chain 1 |
| chr5_+_76957812 | 3.09 |
ENSMUST00000120818.8
|
Cracd
|
capping protein inhibiting regulator of actin |
| chrY_+_57195716 | 3.09 |
ENSMUST00000189109.7
ENSMUST00000191355.7 ENSMUST00000190292.2 |
Sly
|
Sycp3 like Y-linked |
| chr2_+_103953083 | 3.07 |
ENSMUST00000040374.6
|
A930018P22Rik
|
RIKEN cDNA A930018P22 gene |
| chr11_+_70506674 | 3.06 |
ENSMUST00000180052.8
|
4930544D05Rik
|
RIKEN cDNA 4930544D05 gene |
| chr15_-_77330396 | 3.03 |
ENSMUST00000229434.2
|
Apol7b
|
apolipoprotein L 7b |
| chr13_-_92024994 | 3.02 |
ENSMUST00000022122.4
|
Ckmt2
|
creatine kinase, mitochondrial 2 |
| chr18_-_22304435 | 3.01 |
ENSMUST00000234661.2
ENSMUST00000025160.9 |
Ccdc178
|
coiled coil domain containing 178 |
| chr12_+_65272287 | 3.01 |
ENSMUST00000046331.5
ENSMUST00000221658.2 |
Wdr20rt
|
WD repeat domain 20, retrogene |
| chr3_+_96737385 | 3.00 |
ENSMUST00000058865.14
|
Pdzk1
|
PDZ domain containing 1 |
| chrX_+_72760183 | 3.00 |
ENSMUST00000002084.14
|
Abcd1
|
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr4_-_116485118 | 3.00 |
ENSMUST00000030456.14
|
Nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr9_+_106503781 | 3.00 |
ENSMUST00000171091.4
|
Iqcf6
|
IQ motif containing F6 |
| chr12_-_114579938 | 3.00 |
ENSMUST00000195469.6
ENSMUST00000109711.4 |
Ighv1-12
|
immunoglobulin heavy variable V1-12 |
| chr10_-_82152373 | 2.99 |
ENSMUST00000217661.2
|
4932415D10Rik
|
RIKEN cDNA 4932415D10 gene |
| chr6_+_68414401 | 2.99 |
ENSMUST00000103324.3
|
Igkv15-103
|
immunoglobulin kappa chain variable 15-103 |
| chr8_+_27834422 | 2.99 |
ENSMUST00000095375.5
|
Tex24
|
testis expressed gene 24 |
| chr1_-_44118902 | 2.98 |
ENSMUST00000238662.2
|
Gm8251
|
predicted gene 8251 |
| chr9_+_20940669 | 2.98 |
ENSMUST00000001040.7
ENSMUST00000215077.2 |
Icam4
|
intercellular adhesion molecule 4, Landsteiner-Wiener blood group |
| chr1_-_190897012 | 2.98 |
ENSMUST00000171798.2
|
Fam71a
|
family with sequence similarity 71, member A |
| chr18_-_77855446 | 2.95 |
ENSMUST00000048192.9
|
Haus1
|
HAUS augmin-like complex, subunit 1 |
| chr17_+_29712008 | 2.92 |
ENSMUST00000234665.2
|
Pim1
|
proviral integration site 1 |
| chrX_+_85235370 | 2.90 |
ENSMUST00000026036.5
|
Nr0b1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr16_+_32427738 | 2.90 |
ENSMUST00000023486.15
|
Tfrc
|
transferrin receptor |
| chr17_+_9207180 | 2.87 |
ENSMUST00000151609.2
ENSMUST00000232775.2 ENSMUST00000136954.3 |
1700010I14Rik
|
RIKEN cDNA 1700010I14 gene |
| chr5_-_123620632 | 2.86 |
ENSMUST00000198901.2
|
Il31
|
interleukin 31 |
| chr3_+_82933383 | 2.83 |
ENSMUST00000029630.15
ENSMUST00000166581.4 |
Fga
|
fibrinogen alpha chain |
| chrY_+_65386218 | 2.83 |
ENSMUST00000190282.7
ENSMUST00000185550.7 |
Gm20736
|
predicted gene, 20736 |
| chr4_+_136337742 | 2.83 |
ENSMUST00000046647.3
|
Tex46
|
testis expressed 46 |
| chr1_+_160806241 | 2.82 |
ENSMUST00000195760.2
|
Serpinc1
|
serine (or cysteine) peptidase inhibitor, clade C (antithrombin), member 1 |
| chr12_+_4819277 | 2.82 |
ENSMUST00000020967.11
|
Pfn4
|
profilin family, member 4 |
| chr8_-_3767547 | 2.81 |
ENSMUST00000058040.7
|
Clec4g
|
C-type lectin domain family 4, member g |
| chr12_+_65272495 | 2.80 |
ENSMUST00000221980.2
|
Wdr20rt
|
WD repeat domain 20, retrogene |
| chr8_+_84075066 | 2.80 |
ENSMUST00000038692.6
|
Mgat4d
|
MGAT4 family, member C |
| chr18_-_22304459 | 2.80 |
ENSMUST00000234736.2
|
Ccdc178
|
coiled coil domain containing 178 |
| chr12_-_115109539 | 2.80 |
ENSMUST00000192554.6
ENSMUST00000103522.3 |
Ighv1-52
|
immunoglobulin heavy variable 1-52 |
| chr9_-_119812829 | 2.79 |
ENSMUST00000216929.2
|
Csrnp1
|
cysteine-serine-rich nuclear protein 1 |
| chr7_+_43751749 | 2.78 |
ENSMUST00000085455.6
|
Klk1b21
|
kallikrein 1-related peptidase b21 |
| chr18_-_65527078 | 2.77 |
ENSMUST00000035548.16
|
Alpk2
|
alpha-kinase 2 |
| chr17_+_34341766 | 2.77 |
ENSMUST00000042121.11
|
H2-DMa
|
histocompatibility 2, class II, locus DMa |
| chr5_+_122344854 | 2.76 |
ENSMUST00000145854.8
|
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr1_-_192883743 | 2.75 |
ENSMUST00000043550.11
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr12_+_4819339 | 2.75 |
ENSMUST00000178879.3
|
Pfn4
|
profilin family, member 4 |
| chr17_-_45903410 | 2.74 |
ENSMUST00000166119.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr17_-_45903188 | 2.74 |
ENSMUST00000164769.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr7_-_43309563 | 2.73 |
ENSMUST00000032667.10
|
Siglece
|
sialic acid binding Ig-like lectin E |
| chr12_-_114023935 | 2.73 |
ENSMUST00000103469.4
|
Ighv14-3
|
immunoglobulin heavy variable V14-3 |
| chr7_+_44890497 | 2.73 |
ENSMUST00000213347.2
|
Slc6a16
|
solute carrier family 6, member 16 |
| chr6_+_29471436 | 2.72 |
ENSMUST00000171317.2
|
Atp6v1fnb
|
Atp6v1f neighbor |
| chr9_-_119019450 | 2.72 |
ENSMUST00000093775.12
|
Slc22a14
|
solute carrier family 22 (organic cation transporter), member 14 |
| chr18_-_74340842 | 2.71 |
ENSMUST00000040188.16
|
Ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr11_-_102298281 | 2.71 |
ENSMUST00000107098.8
ENSMUST00000018821.9 |
Slc25a39
|
solute carrier family 25, member 39 |
| chrX_-_94521712 | 2.69 |
ENSMUST00000033549.3
|
Asb12
|
ankyrin repeat and SOCS box-containing 12 |
| chr12_-_114955196 | 2.69 |
ENSMUST00000194865.2
|
Ighv1-47
|
immunoglobulin heavy variable 1-47 |
| chr16_+_29028860 | 2.68 |
ENSMUST00000162747.8
|
Plaat1
|
phospholipase A and acyltransferase 1 |
| chr17_-_33358473 | 2.67 |
ENSMUST00000131954.2
ENSMUST00000053896.8 |
Morc2b
|
microrchidia 2B |
| chr13_+_65065220 | 2.67 |
ENSMUST00000070216.9
ENSMUST00000221202.2 |
Spata31
|
spermatogenesis associated 31 |
| chr10_+_79500387 | 2.66 |
ENSMUST00000020554.8
|
Madcam1
|
mucosal vascular addressin cell adhesion molecule 1 |
| chr15_-_20666836 | 2.65 |
ENSMUST00000052910.6
|
Acot10
|
acyl-CoA thioesterase 10 |
| chr2_-_110983281 | 2.65 |
ENSMUST00000132464.2
|
Gm15130
|
predicted gene 15130 |
| chr11_-_59678462 | 2.65 |
ENSMUST00000125307.2
|
Pld6
|
phospholipase D family, member 6 |
| chr18_-_74340885 | 2.64 |
ENSMUST00000177604.2
|
Ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr12_-_114579763 | 2.64 |
ENSMUST00000103500.2
|
Ighv1-12
|
immunoglobulin heavy variable V1-12 |
| chr9_+_110673565 | 2.63 |
ENSMUST00000176403.8
|
Prss46
|
protease, serine 46 |
| chr5_-_134935579 | 2.63 |
ENSMUST00000201316.4
ENSMUST00000047305.6 |
Tmem270
|
transmembrane protein 270 |
| chr6_-_135287372 | 2.62 |
ENSMUST00000050471.4
|
Pbp2
|
phosphatidylethanolamine binding protein 2 |
| chr10_+_61010983 | 2.61 |
ENSMUST00000143207.8
ENSMUST00000148181.8 ENSMUST00000151886.8 |
Tbata
|
thymus, brain and testes associated |
| chr4_+_135639117 | 2.60 |
ENSMUST00000068830.4
|
Cnr2
|
cannabinoid receptor 2 (macrophage) |
| chrX_+_77781343 | 2.58 |
ENSMUST00000105110.4
|
Fam47c
|
family with sequence similarity 47, member C |
| chr10_+_79722081 | 2.58 |
ENSMUST00000046091.7
|
Elane
|
elastase, neutrophil expressed |
| chr3_-_75072319 | 2.58 |
ENSMUST00000124618.2
|
Zbbx
|
zinc finger, B-box domain containing |
| chr9_-_119019428 | 2.57 |
ENSMUST00000127794.2
|
Slc22a14
|
solute carrier family 22 (organic cation transporter), member 14 |
| chr1_+_87111033 | 2.57 |
ENSMUST00000044533.9
|
Prss56
|
protease, serine 56 |
| chr4_-_42756489 | 2.56 |
ENSMUST00000140546.3
ENSMUST00000102957.6 |
Ccl19
|
chemokine (C-C motif) ligand 19 |
| chrX_-_9335525 | 2.54 |
ENSMUST00000015484.10
|
Cybb
|
cytochrome b-245, beta polypeptide |
| chr12_-_114878652 | 2.54 |
ENSMUST00000103515.2
|
Ighv1-39
|
immunoglobulin heavy variable 1-39 |
| chr12_-_114451189 | 2.53 |
ENSMUST00000103493.3
|
Ighv1-4
|
immunoglobulin heavy variable 1-4 |
| chr12_-_114502585 | 2.53 |
ENSMUST00000103496.4
|
Ighv1-7
|
immunoglobulin heavy variable V1-7 |
| chr2_-_35257741 | 2.53 |
ENSMUST00000028243.2
|
4930568D16Rik
|
RIKEN cDNA 4930568D16 gene |
| chr2_+_34999497 | 2.51 |
ENSMUST00000028235.11
ENSMUST00000156933.8 ENSMUST00000028237.15 ENSMUST00000113032.8 |
Cntrl
|
centriolin |
| chr7_-_100160987 | 2.51 |
ENSMUST00000054923.9
|
Dnajb13
|
DnaJ heat shock protein family (Hsp40) member B13 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 20.3 | GO:0019516 | lactate oxidation(GO:0019516) |
| 2.0 | 6.1 | GO:0030221 | basophil differentiation(GO:0030221) |
| 1.7 | 5.1 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 1.6 | 4.8 | GO:0097037 | heme export(GO:0097037) |
| 1.4 | 5.7 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 1.3 | 5.2 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 1.2 | 3.7 | GO:0071846 | actin filament debranching(GO:0071846) |
| 1.2 | 3.7 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) |
| 1.2 | 3.5 | GO:0002215 | defense response to nematode(GO:0002215) |
| 1.2 | 3.5 | GO:0002396 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 1.1 | 7.8 | GO:0015862 | uridine transport(GO:0015862) |
| 1.1 | 4.4 | GO:1902490 | regulation of sperm capacitation(GO:1902490) |
| 1.0 | 5.2 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.9 | 149.8 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.9 | 7.3 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.9 | 2.7 | GO:1903401 | nitric oxide production involved in inflammatory response(GO:0002537) lysine import(GO:0034226) L-lysine import(GO:0061461) L-ornithine transmembrane transport(GO:1903352) L-arginine transmembrane transport(GO:1903400) L-lysine transmembrane transport(GO:1903401) L-lysine import into cell(GO:1903410) arginine transmembrane transport(GO:1903826) |
| 0.9 | 2.7 | GO:0007309 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 0.9 | 2.6 | GO:0002777 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) neutrophil mediated killing of fungus(GO:0070947) |
| 0.9 | 9.4 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.9 | 2.6 | GO:2000547 | regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.8 | 5.9 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.8 | 5.0 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.8 | 3.3 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.8 | 2.3 | GO:0045404 | interleukin-10 biosynthetic process(GO:0042091) interleukin-13 biosynthetic process(GO:0042231) regulation of interleukin-10 biosynthetic process(GO:0045074) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.8 | 6.9 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.8 | 3.8 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.7 | 3.7 | GO:2000520 | regulation of immunological synapse formation(GO:2000520) |
| 0.7 | 3.7 | GO:0006231 | dTMP biosynthetic process(GO:0006231) dTMP metabolic process(GO:0046073) |
| 0.7 | 2.8 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.7 | 3.5 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.7 | 2.1 | GO:1900062 | regulation of replicative cell aging(GO:1900062) |
| 0.7 | 2.7 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) |
| 0.7 | 4.0 | GO:0032824 | negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.7 | 3.3 | GO:1904253 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.6 | 2.5 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.6 | 2.5 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.6 | 4.8 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.6 | 3.6 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.6 | 3.6 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.6 | 4.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.6 | 2.3 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.6 | 8.9 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.6 | 1.7 | GO:0014728 | regulation of the force of skeletal muscle contraction(GO:0014728) regulation of skeletal muscle contraction by chemo-mechanical energy conversion(GO:0014862) regulation of NAD metabolic process(GO:1902688) regulation of glucose catabolic process to lactate via pyruvate(GO:1904023) |
| 0.6 | 3.9 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.6 | 2.2 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.5 | 1.6 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.5 | 1.0 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.5 | 1.5 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.5 | 5.0 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.5 | 2.0 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.5 | 2.0 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.5 | 6.2 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.5 | 1.4 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.4 | 1.3 | GO:1904582 | positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.4 | 4.0 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.4 | 2.2 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.4 | 5.7 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.4 | 6.5 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.4 | 4.2 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.4 | 4.9 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.4 | 2.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.4 | 1.6 | GO:0001905 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.4 | 1.2 | GO:2000813 | actin filament uncapping(GO:0051695) negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.4 | 1.2 | GO:0072302 | metanephric comma-shaped body morphogenesis(GO:0072278) negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 0.4 | 2.3 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.4 | 2.3 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.4 | 2.3 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.4 | 3.4 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.4 | 0.4 | GO:1904173 | regulation of histone demethylase activity (H3-K4 specific)(GO:1904173) |
| 0.4 | 1.5 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.4 | 3.0 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.4 | 3.7 | GO:0045348 | positive regulation of MHC class II biosynthetic process(GO:0045348) |
| 0.4 | 1.5 | GO:0015886 | heme transport(GO:0015886) |
| 0.4 | 1.8 | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.4 | 1.1 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.4 | 2.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.4 | 1.1 | GO:0098583 | thorax and anterior abdomen determination(GO:0007356) mastication(GO:0071626) learned vocalization behavior(GO:0098583) |
| 0.3 | 2.3 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.3 | 2.9 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.3 | 2.6 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.3 | 3.8 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.3 | 4.6 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.3 | 1.8 | GO:0002442 | serotonin production involved in inflammatory response(GO:0002351) serotonin secretion involved in inflammatory response(GO:0002442) serotonin secretion by platelet(GO:0002554) |
| 0.3 | 1.8 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.3 | 4.2 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.3 | 4.7 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.3 | 9.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.3 | 0.9 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.3 | 0.9 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.3 | 1.4 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.3 | 1.7 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.3 | 2.8 | GO:0060051 | negative regulation of protein glycosylation(GO:0060051) |
| 0.3 | 1.7 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.3 | 3.0 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.3 | 3.0 | GO:1902033 | regulation of hematopoietic stem cell proliferation(GO:1902033) |
| 0.3 | 1.3 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.3 | 1.3 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.3 | 0.8 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.3 | 2.9 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.3 | 2.1 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.2 | 1.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.2 | 4.4 | GO:0071281 | cellular response to iron ion(GO:0071281) |
| 0.2 | 0.7 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.2 | 3.1 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.2 | 1.7 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 | 3.6 | GO:0001787 | natural killer cell proliferation(GO:0001787) |
| 0.2 | 0.9 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.2 | 0.9 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.2 | 0.7 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.2 | 0.7 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.2 | 0.9 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 1.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.2 | 2.0 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.2 | 0.7 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.2 | 1.1 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.2 | 2.8 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.2 | 0.4 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.2 | 1.4 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.2 | 8.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.2 | 5.1 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.2 | 1.2 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.2 | 3.3 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.2 | 0.6 | GO:0046436 | D-serine catabolic process(GO:0036088) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.2 | 3.2 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.2 | 2.2 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.2 | 0.9 | GO:1904976 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.2 | 0.4 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.2 | 5.3 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) |
| 0.2 | 3.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 2.3 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.2 | 2.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.2 | 1.2 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.2 | 0.5 | GO:0019740 | regulation of nitrogen utilization(GO:0006808) nitrogen utilization(GO:0019740) |
| 0.2 | 0.5 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.2 | 1.7 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 0.2 | 0.9 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.2 | 77.3 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.2 | 0.5 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.2 | 1.5 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.2 | 4.3 | GO:0080184 | response to phenylpropanoid(GO:0080184) |
| 0.2 | 1.4 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.2 | 0.5 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.2 | 2.8 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.2 | 0.2 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.2 | 0.3 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.2 | 1.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 4.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 1.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 1.0 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 2.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 1.2 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.1 | 1.6 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.1 | 3.7 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 0.7 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.1 | 1.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 0.7 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 2.8 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.1 | 7.0 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 0.5 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 | 0.1 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) |
| 0.1 | 19.6 | GO:0048515 | spermatid differentiation(GO:0048515) |
| 0.1 | 0.5 | GO:0046061 | dGTP catabolic process(GO:0006203) dATP catabolic process(GO:0046061) |
| 0.1 | 1.0 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 1.7 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.5 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.1 | 2.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.4 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.1 | 1.9 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 3.2 | GO:2000637 | positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.1 | 1.4 | GO:1904948 | midbrain dopaminergic neuron differentiation(GO:1904948) |
| 0.1 | 1.4 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.9 | GO:0006956 | complement activation(GO:0006956) |
| 0.1 | 1.4 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 0.8 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 1.0 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 1.7 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 1.7 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 0.3 | GO:0015838 | amino-acid betaine transport(GO:0015838) |
| 0.1 | 1.0 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.1 | 2.4 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 40.7 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.1 | 2.7 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.1 | 1.1 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 2.9 | GO:0043968 | N-terminal protein amino acid acetylation(GO:0006474) histone H2A acetylation(GO:0043968) |
| 0.1 | 1.1 | GO:1901072 | glucosamine-containing compound catabolic process(GO:1901072) |
| 0.1 | 1.0 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.1 | 1.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.6 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.3 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 1.6 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 1.3 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 3.1 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 0.2 | GO:0014064 | positive regulation of serotonin secretion(GO:0014064) |
| 0.1 | 5.0 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 0.3 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 | 0.4 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.1 | 4.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 0.5 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 3.7 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.1 | 5.2 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 2.0 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.1 | 1.8 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 1.9 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 0.7 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 3.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 3.1 | GO:0009072 | aromatic amino acid family metabolic process(GO:0009072) |
| 0.1 | 1.1 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 0.5 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 0.2 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.1 | 2.7 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.1 | 1.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 1.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.1 | 1.0 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 0.2 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 1.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 2.0 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 0.3 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.3 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.1 | 1.4 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 0.5 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 0.5 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.1 | 1.3 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.1 | 0.2 | GO:0061402 | positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 0.1 | 1.7 | GO:0006582 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.1 | 1.0 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 1.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 3.3 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 1.0 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.1 | 2.6 | GO:0003341 | cilium movement(GO:0003341) |
| 0.1 | 0.9 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.1 | 0.2 | GO:0036233 | glycine import(GO:0036233) |
| 0.1 | 0.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 2.1 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.7 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.5 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.5 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.5 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.4 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.0 | 0.4 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 1.3 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.0 | 0.4 | GO:0042640 | anagen(GO:0042640) |
| 0.0 | 1.4 | GO:0001773 | myeloid dendritic cell activation(GO:0001773) |
| 0.0 | 5.1 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.4 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.9 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.3 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.3 | GO:0042424 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.2 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 1.2 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.5 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 | 0.4 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 2.5 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.0 | 1.8 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.0 | 1.4 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.5 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.4 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.3 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 2.1 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.0 | 0.6 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 1.0 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 0.6 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 1.6 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 2.4 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 3.2 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.0 | 0.7 | GO:0071353 | response to interleukin-4(GO:0070670) cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.5 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 1.2 | GO:0050798 | activated T cell proliferation(GO:0050798) |
| 0.0 | 0.4 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.0 | 0.3 | GO:1903025 | regulation of histone H3-K36 methylation(GO:0000414) regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.7 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 1.5 | GO:0070303 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.0 | 0.6 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.0 | 4.2 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.1 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.0 | 0.7 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 | 2.7 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.2 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 5.3 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 0.6 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.3 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.5 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.0 | 0.2 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.4 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:2001271 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.4 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.2 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.2 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.0 | 0.4 | GO:0048753 | melanosome organization(GO:0032438) pigment granule organization(GO:0048753) |
| 0.0 | 0.9 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 2.1 | GO:0003281 | ventricular septum development(GO:0003281) |
| 0.0 | 0.2 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.4 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 0.3 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.1 | GO:0060100 | positive regulation of alkaline phosphatase activity(GO:0010694) positive regulation of phagocytosis, engulfment(GO:0060100) positive regulation of membrane invagination(GO:1905155) |
| 0.0 | 0.6 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.3 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 | 0.6 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.7 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 | 1.0 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.6 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 1.2 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.5 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 1.4 | GO:0007276 | gamete generation(GO:0007276) |
| 0.0 | 0.5 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.6 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.6 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.2 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.4 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.0 | 1.8 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.0 | 0.1 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.4 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.5 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.6 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 0.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.8 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.8 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 1.6 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.1 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 1.0 | GO:1903555 | regulation of tumor necrosis factor production(GO:0032680) regulation of tumor necrosis factor superfamily cytokine production(GO:1903555) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 1.2 | 4.8 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 1.1 | 141.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 1.0 | 3.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.9 | 0.9 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.9 | 3.7 | GO:0008623 | CHRAC(GO:0008623) |
| 0.7 | 5.2 | GO:0019814 | immunoglobulin complex(GO:0019814) |
| 0.7 | 3.4 | GO:0070442 | integrin alphaIIb-beta3 complex(GO:0070442) |
| 0.7 | 3.3 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.7 | 8.0 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.6 | 3.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.6 | 7.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.6 | 2.2 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.5 | 6.8 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.5 | 6.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.5 | 6.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.5 | 71.9 | GO:0000800 | lateral element(GO:0000800) |
| 0.5 | 3.3 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.4 | 3.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.4 | 1.8 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.4 | 5.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.3 | 1.0 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.3 | 2.0 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.3 | 1.3 | GO:0005757 | mitochondrial permeability transition pore complex(GO:0005757) |
| 0.3 | 1.6 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.3 | 2.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 1.2 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.3 | 0.9 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.3 | 9.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.3 | 0.8 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.3 | 1.0 | GO:0032021 | NELF complex(GO:0032021) |
| 0.2 | 7.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 3.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.2 | 1.0 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.2 | 7.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.2 | 4.0 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.2 | 1.0 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.2 | 1.0 | GO:0035363 | histone locus body(GO:0035363) |
| 0.2 | 1.2 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.2 | 5.9 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.2 | 2.0 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 2.0 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.2 | 1.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 1.7 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.2 | 2.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 1.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.2 | 0.7 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.2 | 25.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 0.7 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.2 | 3.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 5.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 2.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 3.0 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 0.9 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 1.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 29.1 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 1.5 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 0.4 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 1.8 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 12.1 | GO:0097014 | ciliary plasm(GO:0097014) |
| 0.1 | 4.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 1.2 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 2.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 2.9 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.4 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 0.6 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 1.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 1.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 5.7 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 1.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 2.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 1.7 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 10.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 1.0 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 1.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 2.5 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 1.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.4 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 2.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 1.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 0.5 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 2.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.1 | 4.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 0.8 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 7.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 6.9 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 0.5 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 4.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.9 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 1.7 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 0.6 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.5 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 1.0 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 1.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 1.2 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.8 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 1.8 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 3.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 2.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.8 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.6 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 2.1 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.6 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 2.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 2.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 1.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.9 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.7 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.2 | GO:0031932 | TORC2 complex(GO:0031932) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 1.8 | 20.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 1.7 | 5.0 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 1.4 | 4.1 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 1.2 | 4.9 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 1.2 | 9.4 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 1.1 | 141.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 1.0 | 5.0 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 1.0 | 6.8 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.9 | 2.7 | GO:0031370 | eukaryotic initiation factor 4G binding(GO:0031370) |
| 0.9 | 2.7 | GO:0005287 | high-affinity basic amino acid transmembrane transporter activity(GO:0005287) high-affinity arginine transmembrane transporter activity(GO:0005289) high-affinity lysine transmembrane transporter activity(GO:0005292) |
| 0.9 | 2.7 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.8 | 3.1 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.7 | 4.0 | GO:0016019 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.6 | 1.9 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.6 | 2.6 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.6 | 1.9 | GO:0019862 | IgA binding(GO:0019862) |
| 0.6 | 3.6 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.6 | 5.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.6 | 1.7 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.6 | 8.9 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.5 | 4.8 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.5 | 2.1 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.5 | 5.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.5 | 3.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.5 | 4.0 | GO:0005534 | galactose binding(GO:0005534) |
| 0.5 | 3.9 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.5 | 1.4 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.5 | 2.8 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.5 | 6.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.4 | 1.7 | GO:0001512 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.4 | 3.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.4 | 2.5 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.4 | 3.3 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.4 | 2.3 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.4 | 1.6 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.4 | 1.9 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.4 | 1.5 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.4 | 8.2 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.4 | 2.5 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.3 | 2.4 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.3 | 1.0 | GO:0042602 | riboflavin reductase (NADPH) activity(GO:0042602) |
| 0.3 | 6.1 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.3 | 1.6 | GO:0002046 | opsin binding(GO:0002046) |
| 0.3 | 4.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.3 | 1.6 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.3 | 4.0 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.3 | 3.0 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.3 | 1.8 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.3 | 2.9 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.3 | 3.8 | GO:0035473 | lipase binding(GO:0035473) |
| 0.3 | 2.9 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.3 | 0.8 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 0.3 | 8.3 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.3 | 4.8 | GO:0015250 | water channel activity(GO:0015250) |
| 0.3 | 1.3 | GO:0055056 | dehydroascorbic acid transporter activity(GO:0033300) D-glucose transmembrane transporter activity(GO:0055056) |
| 0.3 | 1.3 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.3 | 1.6 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.3 | 1.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 2.7 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 1.7 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.2 | 1.0 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.2 | 1.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 0.7 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.2 | 0.7 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 0.2 | 2.3 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.2 | 2.4 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.2 | 2.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 1.5 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.2 | 2.8 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.2 | 2.5 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.2 | 2.5 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.2 | 4.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 1.0 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.2 | 1.3 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.2 | 1.0 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.2 | 1.8 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.2 | 3.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 5.0 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.2 | 3.7 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.2 | 3.5 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.2 | 3.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 1.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 6.1 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.2 | 2.0 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 3.7 | GO:0001848 | complement binding(GO:0001848) |
| 0.2 | 1.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.2 | 1.2 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.2 | 0.5 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.2 | 0.5 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) |
| 0.2 | 0.5 | GO:0004823 | leucine-tRNA ligase activity(GO:0004823) |
| 0.2 | 0.9 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.2 | 8.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 1.2 | GO:0044729 | double-stranded methylated DNA binding(GO:0010385) hemi-methylated DNA-binding(GO:0044729) |
| 0.2 | 1.2 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.2 | 4.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 1.0 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.2 | 2.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 18.0 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.2 | 1.5 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 0.6 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.2 | 7.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.2 | 3.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.2 | 1.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 2.8 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.2 | 1.8 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 1.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.7 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 0.6 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.1 | 1.5 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.1 | 3.5 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 0.5 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 1.8 | GO:0046977 | TAP binding(GO:0046977) |
| 0.1 | 0.5 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 0.1 | 1.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 1.2 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.1 | 2.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 2.3 | GO:0030547 | receptor inhibitor activity(GO:0030547) |
| 0.1 | 3.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 4.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 0.4 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.1 | 0.3 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 1.8 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 2.9 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 0.5 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 2.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 1.0 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 2.9 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.1 | 2.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.3 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.1 | 0.3 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 1.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.7 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 6.8 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 3.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.5 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.1 | 0.3 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.1 | 0.6 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 1.0 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 4.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.3 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.1 | 0.4 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 6.8 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 0.5 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 7.8 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 1.0 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 2.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 2.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 1.6 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 1.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.4 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 0.8 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 0.2 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 0.1 | 0.2 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 1.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 1.7 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 0.2 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 0.4 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 0.5 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 1.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.3 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 1.0 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 6.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.9 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.5 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 2.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 1.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 3.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 2.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.4 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.4 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 7.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 6.1 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 0.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 2.5 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 4.8 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.0 | 0.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.4 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 2.7 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 0.3 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 1.0 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.4 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 3.5 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 4.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.2 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 5.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.6 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 1.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 4.7 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 0.5 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.5 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.8 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.2 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 2.8 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 2.1 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.1 | GO:0098634 | protein binding involved in cell-matrix adhesion(GO:0098634) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.4 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.6 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.5 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 1.2 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.5 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 2.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.4 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.5 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 1.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.3 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.0 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 11.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 2.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 2.9 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 10.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 6.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 2.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 4.0 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 4.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 3.1 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 1.9 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 3.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 3.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 1.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 4.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 7.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 3.0 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 2.0 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 2.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 5.2 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 2.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 2.7 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 3.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 4.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 1.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.1 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 2.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.6 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 2.5 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 4.5 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.6 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.4 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 5.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 1.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 12.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.7 | 5.6 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.6 | 5.2 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.6 | 6.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.5 | 7.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.4 | 7.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.3 | 4.8 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 3.1 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.3 | 3.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.3 | 4.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 9.1 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.2 | 1.8 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.2 | 3.3 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.2 | 9.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 1.8 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.2 | 3.9 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 2.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 7.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 2.1 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 2.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 7.8 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 5.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 5.5 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 3.6 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.1 | 4.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.2 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 2.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 2.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.0 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 7.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.6 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 0.9 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 2.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 3.3 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 0.9 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 3.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.7 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 3.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 0.7 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.5 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 1.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.0 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.0 | 2.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 1.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.7 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 1.9 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 1.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 2.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.7 | REACTOME FORMATION OF RNA POL II ELONGATION COMPLEX | Genes involved in Formation of RNA Pol II elongation complex |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 2.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |