Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
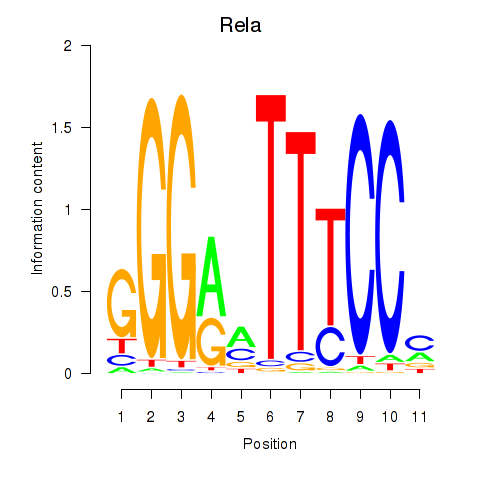
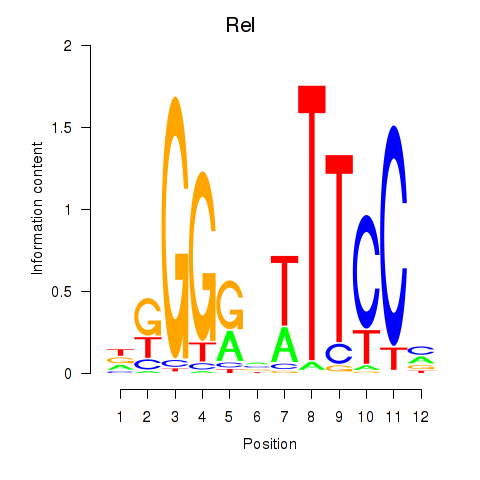
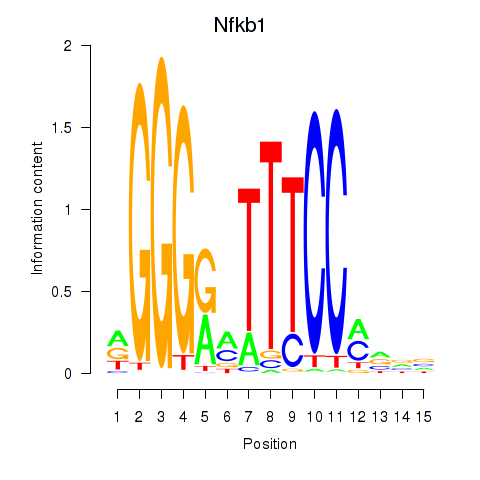
Results for Rela_Rel_Nfkb1
Z-value: 2.88
Transcription factors associated with Rela_Rel_Nfkb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rela
|
ENSMUSG00000024927.9 | Rela |
|
Rel
|
ENSMUSG00000020275.10 | Rel |
|
Nfkb1
|
ENSMUSG00000028163.18 | Nfkb1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfkb1 | mm39_v1_chr3_-_135397298_135397309 | 0.83 | 2.6e-19 | Click! |
| Rel | mm39_v1_chr11_-_23720953_23721028 | 0.62 | 6.9e-09 | Click! |
| Rela | mm39_v1_chr19_+_5687503_5687514 | 0.45 | 8.2e-05 | Click! |
Activity profile of Rela_Rel_Nfkb1 motif
Sorted Z-values of Rela_Rel_Nfkb1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Rela_Rel_Nfkb1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_60936910 | 48.92 |
ENSMUST00000097563.9
ENSMUST00000050487.16 ENSMUST00000167610.2 |
Cd74
|
CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated) |
| chr6_-_129252396 | 37.67 |
ENSMUST00000032259.6
|
Cd69
|
CD69 antigen |
| chr17_+_37504783 | 37.55 |
ENSMUST00000038844.7
|
Ubd
|
ubiquitin D |
| chr17_+_35658131 | 33.14 |
ENSMUST00000071951.14
ENSMUST00000116598.10 ENSMUST00000078205.14 ENSMUST00000076256.8 |
H2-Q7
|
histocompatibility 2, Q region locus 7 |
| chr17_+_35413415 | 29.78 |
ENSMUST00000025262.6
ENSMUST00000173600.2 |
Ltb
|
lymphotoxin B |
| chr19_+_46293210 | 27.87 |
ENSMUST00000236591.2
|
Nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 |
| chr9_-_7873017 | 25.99 |
ENSMUST00000013949.15
|
Birc3
|
baculoviral IAP repeat-containing 3 |
| chr11_-_83421333 | 24.71 |
ENSMUST00000035938.3
|
Ccl5
|
chemokine (C-C motif) ligand 5 |
| chr9_-_7873171 | 24.58 |
ENSMUST00000159323.2
ENSMUST00000115673.3 |
Birc3
|
baculoviral IAP repeat-containing 3 |
| chr10_-_18891095 | 24.25 |
ENSMUST00000019997.11
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr19_+_46293160 | 24.17 |
ENSMUST00000073116.13
|
Nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 |
| chr7_-_19363280 | 24.00 |
ENSMUST00000094762.10
ENSMUST00000049912.15 ENSMUST00000098754.5 |
Relb
|
avian reticuloendotheliosis viral (v-rel) oncogene related B |
| chr10_+_79500387 | 23.14 |
ENSMUST00000020554.8
|
Madcam1
|
mucosal vascular addressin cell adhesion molecule 1 |
| chr6_-_129252323 | 20.95 |
ENSMUST00000204411.2
|
Cd69
|
CD69 antigen |
| chr17_+_31427023 | 20.91 |
ENSMUST00000173776.2
ENSMUST00000048656.15 |
Ubash3a
|
ubiquitin associated and SH3 domain containing, A |
| chr17_-_34506744 | 20.12 |
ENSMUST00000174751.2
ENSMUST00000040655.14 |
H2-Aa
|
histocompatibility 2, class II antigen A, alpha |
| chr10_+_79500421 | 19.48 |
ENSMUST00000217748.2
|
Madcam1
|
mucosal vascular addressin cell adhesion molecule 1 |
| chr9_-_7872970 | 19.25 |
ENSMUST00000115672.2
|
Birc3
|
baculoviral IAP repeat-containing 3 |
| chr12_+_85733415 | 19.00 |
ENSMUST00000040536.6
|
Batf
|
basic leucine zipper transcription factor, ATF-like |
| chr1_+_172327569 | 18.86 |
ENSMUST00000111230.8
|
Tagln2
|
transgelin 2 |
| chr1_+_172327812 | 18.85 |
ENSMUST00000192460.2
|
Tagln2
|
transgelin 2 |
| chr12_-_55539372 | 18.79 |
ENSMUST00000021413.9
|
Nfkbia
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, alpha |
| chr12_+_111383864 | 18.08 |
ENSMUST00000220537.2
ENSMUST00000223050.2 ENSMUST00000072646.8 ENSMUST00000223431.2 ENSMUST00000221144.2 ENSMUST00000222437.2 |
Exoc3l4
|
exocyst complex component 3-like 4 |
| chr16_-_55659194 | 17.87 |
ENSMUST00000096026.9
ENSMUST00000036273.13 ENSMUST00000114457.8 |
Nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta |
| chr5_-_92496730 | 17.25 |
ENSMUST00000038816.13
ENSMUST00000118006.3 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr9_-_44437694 | 16.04 |
ENSMUST00000062215.8
|
Cxcr5
|
chemokine (C-X-C motif) receptor 5 |
| chr14_-_55828511 | 16.00 |
ENSMUST00000161807.8
ENSMUST00000111378.10 ENSMUST00000159687.2 |
Psme2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr1_-_135186176 | 15.86 |
ENSMUST00000185752.2
ENSMUST00000003135.14 |
Elf3
|
E74-like factor 3 |
| chr1_-_172418058 | 15.80 |
ENSMUST00000065679.8
|
Slamf8
|
SLAM family member 8 |
| chr11_+_100751272 | 15.30 |
ENSMUST00000107357.4
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chr11_+_100751151 | 15.03 |
ENSMUST00000138083.8
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chr1_+_192984278 | 14.49 |
ENSMUST00000016315.16
|
Lamb3
|
laminin, beta 3 |
| chrX_-_73289970 | 14.30 |
ENSMUST00000130007.8
|
Flna
|
filamin, alpha |
| chr7_+_30121776 | 14.19 |
ENSMUST00000108175.8
|
Nfkbid
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, delta |
| chr10_-_12839995 | 14.18 |
ENSMUST00000219727.2
ENSMUST00000163425.9 ENSMUST00000042861.7 ENSMUST00000218685.2 |
Stx11
|
syntaxin 11 |
| chr19_+_46294119 | 13.99 |
ENSMUST00000111881.4
|
Nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 |
| chr5_+_136996713 | 13.97 |
ENSMUST00000001790.6
|
Cldn15
|
claudin 15 |
| chr3_-_83749036 | 13.74 |
ENSMUST00000029623.11
|
Tlr2
|
toll-like receptor 2 |
| chrX_-_73290140 | 13.71 |
ENSMUST00000101454.9
ENSMUST00000033699.13 |
Flna
|
filamin, alpha |
| chr17_-_35424281 | 13.53 |
ENSMUST00000025266.6
|
Lta
|
lymphotoxin A |
| chr5_+_43976218 | 13.42 |
ENSMUST00000101237.8
|
Bst1
|
bone marrow stromal cell antigen 1 |
| chr17_+_35598583 | 13.20 |
ENSMUST00000081435.5
|
H2-Q4
|
histocompatibility 2, Q region locus 4 |
| chr12_-_114117264 | 13.20 |
ENSMUST00000103461.5
|
Ighv7-3
|
immunoglobulin heavy variable 7-3 |
| chr3_-_135397298 | 12.87 |
ENSMUST00000029812.14
|
Nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 1, p105 |
| chrX_+_7523499 | 12.64 |
ENSMUST00000033485.14
|
Prickle3
|
prickle planar cell polarity protein 3 |
| chr11_+_69471185 | 12.61 |
ENSMUST00000171247.8
ENSMUST00000108658.10 ENSMUST00000005371.12 |
Trp53
|
transformation related protein 53 |
| chr9_+_32607301 | 12.58 |
ENSMUST00000034534.13
ENSMUST00000050797.14 ENSMUST00000184887.2 |
Ets1
|
E26 avian leukemia oncogene 1, 5' domain |
| chr9_+_5308828 | 12.45 |
ENSMUST00000162846.8
ENSMUST00000027012.14 |
Casp4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr11_+_100750316 | 12.35 |
ENSMUST00000107356.8
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chr18_-_4352944 | 11.99 |
ENSMUST00000025078.10
|
Map3k8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr11_+_69471219 | 11.89 |
ENSMUST00000108657.4
|
Trp53
|
transformation related protein 53 |
| chr12_+_79177523 | 11.79 |
ENSMUST00000021550.7
|
Arg2
|
arginase type II |
| chr7_-_126613707 | 11.37 |
ENSMUST00000165096.9
|
Mvp
|
major vault protein |
| chr6_-_69792108 | 11.31 |
ENSMUST00000103367.3
|
Igkv12-44
|
immunoglobulin kappa variable 12-44 |
| chr6_+_68233361 | 11.31 |
ENSMUST00000103320.3
|
Igkv14-111
|
immunoglobulin kappa variable 14-111 |
| chr17_-_23964807 | 11.22 |
ENSMUST00000046525.10
|
Kremen2
|
kringle containing transmembrane protein 2 |
| chr18_+_4994600 | 10.94 |
ENSMUST00000140448.8
|
Svil
|
supervillin |
| chr14_-_30329765 | 10.65 |
ENSMUST00000112207.8
ENSMUST00000112206.8 ENSMUST00000112202.8 ENSMUST00000112203.2 |
Prkcd
|
protein kinase C, delta |
| chr3_-_65865656 | 10.61 |
ENSMUST00000029416.14
|
Ccnl1
|
cyclin L1 |
| chr8_-_62355690 | 10.61 |
ENSMUST00000121785.9
ENSMUST00000034057.14 |
Palld
|
palladin, cytoskeletal associated protein |
| chr11_-_54853729 | 10.43 |
ENSMUST00000108885.8
ENSMUST00000102730.9 ENSMUST00000018482.13 ENSMUST00000108886.8 ENSMUST00000102731.8 |
Tnip1
|
TNFAIP3 interacting protein 1 |
| chr17_-_37794434 | 10.42 |
ENSMUST00000016427.11
ENSMUST00000171139.3 |
H2-M2
|
histocompatibility 2, M region locus 2 |
| chr8_-_25591737 | 10.33 |
ENSMUST00000098866.11
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr6_+_67873135 | 10.28 |
ENSMUST00000103310.3
|
Igkv14-126
|
immunoglobulin kappa variable 14-126 |
| chr1_+_130728639 | 10.24 |
ENSMUST00000112477.9
ENSMUST00000027670.4 |
Fcamr
|
Fc receptor, IgA, IgM, high affinity |
| chr7_+_30121147 | 10.22 |
ENSMUST00000108176.9
|
Nfkbid
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, delta |
| chr2_-_32277773 | 10.14 |
ENSMUST00000050785.14
|
Lcn2
|
lipocalin 2 |
| chr8_-_105357941 | 10.10 |
ENSMUST00000034351.8
|
Rrad
|
Ras-related associated with diabetes |
| chr1_+_135060994 | 10.06 |
ENSMUST00000167080.3
|
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr6_-_68812291 | 10.03 |
ENSMUST00000199143.5
ENSMUST00000103335.3 |
Igkv12-89
|
immunoglobulin kappa chain variable 12-89 |
| chr8_-_25592001 | 9.96 |
ENSMUST00000128715.8
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr7_+_75105282 | 9.93 |
ENSMUST00000207750.2
ENSMUST00000166315.7 |
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr9_-_44920899 | 9.80 |
ENSMUST00000102832.3
|
Cd3e
|
CD3 antigen, epsilon polypeptide |
| chr9_-_44437801 | 9.74 |
ENSMUST00000215661.2
|
Cxcr5
|
chemokine (C-X-C motif) receptor 5 |
| chr3_+_14706781 | 9.51 |
ENSMUST00000029071.9
|
Car13
|
carbonic anhydrase 13 |
| chr1_+_132996237 | 9.41 |
ENSMUST00000239467.2
|
Pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
| chr1_+_64571942 | 9.35 |
ENSMUST00000171164.8
ENSMUST00000187811.7 ENSMUST00000049932.12 ENSMUST00000087366.11 |
Creb1
|
cAMP responsive element binding protein 1 |
| chr1_+_64571768 | 9.34 |
ENSMUST00000185594.7
|
Creb1
|
cAMP responsive element binding protein 1 |
| chr7_-_44180700 | 9.30 |
ENSMUST00000205506.2
|
Spib
|
Spi-B transcription factor (Spi-1/PU.1 related) |
| chr1_-_36312482 | 9.24 |
ENSMUST00000056946.8
|
Neurl3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr8_-_25592385 | 9.22 |
ENSMUST00000064883.14
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr17_+_35643818 | 9.14 |
ENSMUST00000174699.8
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr7_-_25488060 | 9.11 |
ENSMUST00000002677.11
ENSMUST00000085948.11 |
Axl
|
AXL receptor tyrosine kinase |
| chr6_-_69835868 | 9.11 |
ENSMUST00000103369.2
|
Igkv12-41
|
immunoglobulin kappa chain variable 12-41 |
| chr17_+_35643853 | 9.06 |
ENSMUST00000113879.4
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr10_-_79940168 | 9.04 |
ENSMUST00000219260.2
|
Sbno2
|
strawberry notch 2 |
| chr12_-_84240781 | 9.02 |
ENSMUST00000110294.2
|
Mideas
|
mitotic deacetylase associated SANT domain protein |
| chr11_-_54846873 | 8.99 |
ENSMUST00000155316.2
ENSMUST00000108889.10 ENSMUST00000126703.8 |
Tnip1
|
TNFAIP3 interacting protein 1 |
| chr11_+_117700479 | 8.79 |
ENSMUST00000026649.14
ENSMUST00000177131.8 ENSMUST00000120928.2 ENSMUST00000175737.2 ENSMUST00000132298.2 |
Syngr2
Gm20708
|
synaptogyrin 2 predicted gene 20708 |
| chr18_-_35855383 | 8.58 |
ENSMUST00000133064.8
|
Ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr12_-_115471634 | 8.47 |
ENSMUST00000103535.3
|
Ighv1-64
|
immunoglobulin heavy variable 1-64 |
| chr5_-_105287405 | 8.31 |
ENSMUST00000100961.5
ENSMUST00000031235.13 ENSMUST00000197799.2 ENSMUST00000199629.2 ENSMUST00000196677.5 ENSMUST00000100962.8 |
Gbp9
Gbp8
Gbp4
|
guanylate-binding protein 9 guanylate-binding protein 8 guanylate binding protein 4 |
| chr6_+_48818410 | 8.23 |
ENSMUST00000101426.11
ENSMUST00000168406.4 |
Tmem176a
|
transmembrane protein 176A |
| chr11_-_23720953 | 8.20 |
ENSMUST00000102864.5
|
Rel
|
reticuloendotheliosis oncogene |
| chr8_-_89362745 | 8.19 |
ENSMUST00000034087.9
|
Snx20
|
sorting nexin 20 |
| chr5_-_105441554 | 8.06 |
ENSMUST00000050011.10
ENSMUST00000196520.2 |
Gm43302
Gbp6
|
predicted gene 43302 guanylate binding protein 6 |
| chr1_-_134006847 | 7.79 |
ENSMUST00000020692.7
|
Btg2
|
BTG anti-proliferation factor 2 |
| chr14_+_54457978 | 7.78 |
ENSMUST00000103740.2
ENSMUST00000198398.5 |
Trac
|
T cell receptor alpha constant |
| chr7_+_24069680 | 7.78 |
ENSMUST00000205428.2
ENSMUST00000171904.3 ENSMUST00000205626.2 |
Kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr1_+_64572050 | 7.77 |
ENSMUST00000190348.2
|
Creb1
|
cAMP responsive element binding protein 1 |
| chr14_+_80237691 | 7.75 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr8_+_47192911 | 7.62 |
ENSMUST00000208433.2
|
Irf2
|
interferon regulatory factor 2 |
| chr4_+_129713991 | 7.62 |
ENSMUST00000030578.14
|
Ptp4a2
|
protein tyrosine phosphatase 4a2 |
| chr14_-_54655079 | 7.57 |
ENSMUST00000226753.2
ENSMUST00000197440.5 |
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr6_-_48817914 | 7.50 |
ENSMUST00000164733.4
|
Tmem176b
|
transmembrane protein 176B |
| chr6_+_41090484 | 7.50 |
ENSMUST00000103267.3
|
Trbv12-1
|
T cell receptor beta, variable 12-1 |
| chr1_+_159871943 | 7.43 |
ENSMUST00000163892.8
|
4930523C07Rik
|
RIKEN cDNA 4930523C07 gene |
| chr8_-_105357872 | 7.42 |
ENSMUST00000212163.2
|
Rrad
|
Ras-related associated with diabetes |
| chr12_-_113823290 | 7.42 |
ENSMUST00000103459.5
|
Ighv5-17
|
immunoglobulin heavy variable 5-17 |
| chr12_+_111409087 | 7.35 |
ENSMUST00000109792.8
|
Tnfaip2
|
tumor necrosis factor, alpha-induced protein 2 |
| chr17_+_56259617 | 7.32 |
ENSMUST00000003274.8
|
Ebi3
|
Epstein-Barr virus induced gene 3 |
| chr7_-_126651847 | 7.28 |
ENSMUST00000205424.2
|
Zg16
|
zymogen granule protein 16 |
| chr15_-_76501525 | 7.24 |
ENSMUST00000230977.2
|
Slc39a4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr11_-_69771797 | 7.23 |
ENSMUST00000238978.2
|
Kctd11
|
potassium channel tetramerisation domain containing 11 |
| chr4_+_130643260 | 7.23 |
ENSMUST00000030316.7
|
Laptm5
|
lysosomal-associated protein transmembrane 5 |
| chr11_-_99045894 | 7.13 |
ENSMUST00000103134.4
|
Ccr7
|
chemokine (C-C motif) receptor 7 |
| chr6_-_78445846 | 7.12 |
ENSMUST00000032089.3
|
Reg3g
|
regenerating islet-derived 3 gamma |
| chr9_-_31043076 | 7.12 |
ENSMUST00000034478.3
|
St14
|
suppression of tumorigenicity 14 (colon carcinoma) |
| chr2_-_118377500 | 7.09 |
ENSMUST00000125860.3
|
Bmf
|
BCL2 modifying factor |
| chr9_+_123596276 | 7.06 |
ENSMUST00000166236.9
ENSMUST00000111454.4 ENSMUST00000168910.2 |
Ccr9
|
chemokine (C-C motif) receptor 9 |
| chr6_-_48818302 | 7.03 |
ENSMUST00000203355.3
ENSMUST00000166247.8 |
Tmem176b
|
transmembrane protein 176B |
| chr11_-_70213348 | 7.01 |
ENSMUST00000019051.3
|
Alox12e
|
arachidonate lipoxygenase, epidermal |
| chr13_+_43938251 | 6.98 |
ENSMUST00000015540.4
|
Cd83
|
CD83 antigen |
| chr3_+_142202642 | 6.97 |
ENSMUST00000090127.7
|
Gbp5
|
guanylate binding protein 5 |
| chr11_-_83469446 | 6.92 |
ENSMUST00000019266.6
|
Ccl9
|
chemokine (C-C motif) ligand 9 |
| chr11_+_120333470 | 6.89 |
ENSMUST00000044105.9
|
Tspan10
|
tetraspanin 10 |
| chr6_-_48818006 | 6.86 |
ENSMUST00000203229.3
|
Tmem176b
|
transmembrane protein 176B |
| chr4_+_151012375 | 6.78 |
ENSMUST00000139826.8
ENSMUST00000116257.8 |
Tnfrsf9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr6_+_41098273 | 6.55 |
ENSMUST00000103270.4
|
Trbv13-2
|
T cell receptor beta, variable 13-2 |
| chr15_+_79779218 | 6.53 |
ENSMUST00000023054.14
|
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr15_+_80456775 | 6.48 |
ENSMUST00000229980.2
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr6_-_48818430 | 6.39 |
ENSMUST00000205147.3
|
Tmem176b
|
transmembrane protein 176B |
| chr17_-_84495364 | 6.37 |
ENSMUST00000060366.7
|
Zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr2_-_32278245 | 6.36 |
ENSMUST00000192241.2
|
Lcn2
|
lipocalin 2 |
| chr4_-_101122433 | 6.34 |
ENSMUST00000149297.2
ENSMUST00000102781.10 |
Jak1
|
Janus kinase 1 |
| chr4_-_125021610 | 6.27 |
ENSMUST00000036188.8
|
Zc3h12a
|
zinc finger CCCH type containing 12A |
| chr1_+_192835414 | 6.25 |
ENSMUST00000076521.7
|
Irf6
|
interferon regulatory factor 6 |
| chr18_-_20192535 | 6.19 |
ENSMUST00000075214.9
ENSMUST00000039247.11 |
Dsc2
|
desmocollin 2 |
| chr7_+_16043502 | 6.16 |
ENSMUST00000002152.13
|
Bbc3
|
BCL2 binding component 3 |
| chr8_-_123159663 | 6.15 |
ENSMUST00000017604.10
|
Cyba
|
cytochrome b-245, alpha polypeptide |
| chr6_-_48818044 | 6.10 |
ENSMUST00000101429.11
ENSMUST00000204073.3 |
Tmem176b
|
transmembrane protein 176B |
| chr1_+_83094481 | 6.08 |
ENSMUST00000027351.13
ENSMUST00000113437.9 ENSMUST00000186832.2 |
Ccl20
|
chemokine (C-C motif) ligand 20 |
| chr11_-_48884999 | 5.98 |
ENSMUST00000146439.8
|
Tgtp1
|
T cell specific GTPase 1 |
| chr10_+_80765900 | 5.96 |
ENSMUST00000015456.10
ENSMUST00000220246.2 |
Gadd45b
|
growth arrest and DNA-damage-inducible 45 beta |
| chr5_-_92475927 | 5.90 |
ENSMUST00000113093.5
|
Cxcl9
|
chemokine (C-X-C motif) ligand 9 |
| chr19_+_34268053 | 5.89 |
ENSMUST00000025691.13
|
Fas
|
Fas (TNF receptor superfamily member 6) |
| chr11_+_83007635 | 5.87 |
ENSMUST00000037994.8
|
Slfn1
|
schlafen 1 |
| chr2_-_103133503 | 5.86 |
ENSMUST00000111176.9
|
Ehf
|
ets homologous factor |
| chr19_+_34268071 | 5.85 |
ENSMUST00000112472.4
ENSMUST00000235232.2 |
Fas
|
Fas (TNF receptor superfamily member 6) |
| chr5_-_18040713 | 5.84 |
ENSMUST00000197890.5
|
Cd36
|
CD36 molecule |
| chr12_-_80307110 | 5.82 |
ENSMUST00000021554.16
|
Actn1
|
actinin, alpha 1 |
| chr17_-_34219225 | 5.77 |
ENSMUST00000238098.2
ENSMUST00000087189.7 ENSMUST00000173075.3 ENSMUST00000172912.8 ENSMUST00000236740.2 ENSMUST00000025181.18 |
H2-K1
|
histocompatibility 2, K1, K region |
| chr17_+_35482063 | 5.75 |
ENSMUST00000172503.3
|
H2-D1
|
histocompatibility 2, D region locus 1 |
| chr1_-_173707677 | 5.74 |
ENSMUST00000190651.4
ENSMUST00000188804.7 |
Mndal
|
myeloid nuclear differentiation antigen like |
| chrX_-_74174450 | 5.72 |
ENSMUST00000114092.8
ENSMUST00000132501.8 ENSMUST00000153318.8 ENSMUST00000155742.2 |
Mpp1
|
membrane protein, palmitoylated |
| chr17_+_35481702 | 5.72 |
ENSMUST00000172785.8
|
H2-D1
|
histocompatibility 2, D region locus 1 |
| chr17_+_37209002 | 5.71 |
ENSMUST00000078438.5
|
Trim31
|
tripartite motif-containing 31 |
| chr13_+_42205491 | 5.68 |
ENSMUST00000060148.6
|
Hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr5_-_18040647 | 5.60 |
ENSMUST00000082367.13
|
Cd36
|
CD36 molecule |
| chr14_+_56122404 | 5.57 |
ENSMUST00000022831.5
|
Khnyn
|
KH and NYN domain containing |
| chr2_+_13578738 | 5.54 |
ENSMUST00000141365.3
ENSMUST00000028062.8 |
Vim
|
vimentin |
| chr10_+_58230203 | 5.53 |
ENSMUST00000105468.2
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr17_+_34406523 | 5.50 |
ENSMUST00000170086.8
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr8_-_123159639 | 5.50 |
ENSMUST00000212600.2
|
Cyba
|
cytochrome b-245, alpha polypeptide |
| chr11_+_48977852 | 5.43 |
ENSMUST00000046704.7
ENSMUST00000203810.3 ENSMUST00000203149.3 |
Ifi47
Olfr56
|
interferon gamma inducible protein 47 olfactory receptor 56 |
| chr9_+_69361348 | 5.43 |
ENSMUST00000134907.8
|
Anxa2
|
annexin A2 |
| chr4_+_43957677 | 5.39 |
ENSMUST00000107855.2
|
Glipr2
|
GLI pathogenesis-related 2 |
| chr17_+_45997248 | 5.37 |
ENSMUST00000024734.8
|
Mrpl14
|
mitochondrial ribosomal protein L14 |
| chr2_-_60711706 | 5.33 |
ENSMUST00000164147.8
ENSMUST00000112509.2 |
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr4_+_43957401 | 5.28 |
ENSMUST00000030202.14
|
Glipr2
|
GLI pathogenesis-related 2 |
| chrX_-_74174608 | 5.27 |
ENSMUST00000033775.9
|
Mpp1
|
membrane protein, palmitoylated |
| chr1_+_135060431 | 5.25 |
ENSMUST00000187985.7
ENSMUST00000049449.11 |
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr3_+_127584251 | 5.25 |
ENSMUST00000164447.3
|
Tifa
|
TRAF-interacting protein with forkhead-associated domain |
| chr14_-_30348153 | 5.23 |
ENSMUST00000112211.9
ENSMUST00000112210.11 |
Prkcd
|
protein kinase C, delta |
| chrX_+_41591476 | 5.22 |
ENSMUST00000115070.8
ENSMUST00000153948.2 |
Sh2d1a
|
SH2 domain containing 1A |
| chr2_-_103133524 | 5.20 |
ENSMUST00000090475.10
|
Ehf
|
ets homologous factor |
| chr7_+_101032021 | 5.19 |
ENSMUST00000141083.9
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr12_-_84265609 | 5.19 |
ENSMUST00000046266.13
ENSMUST00000220974.2 |
Mideas
|
mitotic deacetylase associated SANT domain protein |
| chr4_+_112470794 | 5.16 |
ENSMUST00000058791.14
ENSMUST00000186969.2 |
Skint2
|
selection and upkeep of intraepithelial T cells 2 |
| chr9_+_69360902 | 5.14 |
ENSMUST00000034756.15
ENSMUST00000123470.8 |
Anxa2
|
annexin A2 |
| chr15_+_52575796 | 5.12 |
ENSMUST00000037115.9
|
Med30
|
mediator complex subunit 30 |
| chr3_+_127584449 | 5.10 |
ENSMUST00000171621.3
|
Tifa
|
TRAF-interacting protein with forkhead-associated domain |
| chr12_+_104180795 | 5.09 |
ENSMUST00000121337.8
ENSMUST00000167049.8 ENSMUST00000101080.2 |
Serpina3f
|
serine (or cysteine) peptidase inhibitor, clade A, member 3F |
| chr10_-_18887701 | 5.08 |
ENSMUST00000105527.2
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr16_+_91203123 | 5.08 |
ENSMUST00000023691.12
|
Il10rb
|
interleukin 10 receptor, beta |
| chr5_-_137145030 | 5.06 |
ENSMUST00000054384.6
ENSMUST00000152207.2 |
Trim56
|
tripartite motif-containing 56 |
| chr18_-_32692967 | 4.98 |
ENSMUST00000174000.2
ENSMUST00000174459.2 |
Gypc
|
glycophorin C |
| chr2_+_13579092 | 4.98 |
ENSMUST00000193675.2
|
Vim
|
vimentin |
| chr17_+_34406762 | 4.96 |
ENSMUST00000041633.15
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr11_+_48977888 | 4.89 |
ENSMUST00000214804.2
|
Ifi47
|
interferon gamma inducible protein 47 |
| chr3_+_104696108 | 4.86 |
ENSMUST00000002303.12
|
Rhoc
|
ras homolog family member C |
| chrX_-_74174524 | 4.82 |
ENSMUST00000114091.8
|
Mpp1
|
membrane protein, palmitoylated |
| chr14_-_55344004 | 4.82 |
ENSMUST00000036041.15
|
Ap1g2
|
adaptor protein complex AP-1, gamma 2 subunit |
| chr3_+_86131970 | 4.79 |
ENSMUST00000192145.6
ENSMUST00000194759.6 ENSMUST00000107635.7 |
Lrba
|
LPS-responsive beige-like anchor |
| chr1_+_58834532 | 4.76 |
ENSMUST00000027189.15
|
Casp8
|
caspase 8 |
| chr15_+_6599001 | 4.65 |
ENSMUST00000227175.2
|
Fyb
|
FYN binding protein |
| chr8_-_11329656 | 4.62 |
ENSMUST00000208095.2
|
Col4a1
|
collagen, type IV, alpha 1 |
| chr7_-_109215754 | 4.61 |
ENSMUST00000084738.5
|
Denn2b
|
DENN domain containing 2B |
| chr1_-_167112784 | 4.61 |
ENSMUST00000053686.9
|
Uck2
|
uridine-cytidine kinase 2 |
| chr3_-_101743539 | 4.59 |
ENSMUST00000061831.11
|
Mab21l3
|
mab-21-like 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.1 | 70.4 | GO:0002268 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 10.7 | 42.7 | GO:0060376 | positive regulation of mast cell differentiation(GO:0060376) |
| 9.8 | 48.9 | GO:0002906 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 9.8 | 29.3 | GO:0002632 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 9.3 | 28.0 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 8.8 | 26.5 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 7.1 | 21.3 | GO:1902689 | negative regulation of NAD metabolic process(GO:1902689) negative regulation of glucose catabolic process to lactate via pyruvate(GO:1904024) |
| 6.8 | 13.5 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 6.2 | 24.7 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 5.5 | 16.5 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 5.3 | 15.8 | GO:1902623 | negative regulation of neutrophil migration(GO:1902623) |
| 4.7 | 38.0 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 4.7 | 18.8 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 4.6 | 13.7 | GO:0070340 | positive regulation of interleukin-18 production(GO:0032741) detection of triacyl bacterial lipopeptide(GO:0042495) response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) detection of bacterial lipopeptide(GO:0070340) |
| 4.3 | 17.2 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 4.3 | 17.2 | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 4.3 | 12.9 | GO:1904629 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) response to diterpene(GO:1904629) cellular response to diterpene(GO:1904630) response to glucoside(GO:1904631) cellular response to glucoside(GO:1904632) |
| 4.0 | 15.9 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 4.0 | 71.3 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 3.9 | 11.7 | GO:0031104 | dendrite regeneration(GO:0031104) |
| 3.9 | 11.7 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 3.8 | 11.3 | GO:1904093 | regulation of autophagic cell death(GO:1904092) negative regulation of autophagic cell death(GO:1904093) |
| 3.6 | 47.2 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 3.6 | 7.1 | GO:2000412 | positive regulation of thymocyte migration(GO:2000412) |
| 3.5 | 21.3 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 3.5 | 31.7 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 3.5 | 10.5 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 3.4 | 30.6 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 3.2 | 9.6 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 3.1 | 37.6 | GO:0070842 | aggresome assembly(GO:0070842) |
| 3.0 | 6.1 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 2.4 | 12.2 | GO:0097048 | dendritic cell apoptotic process(GO:0097048) regulation of dendritic cell apoptotic process(GO:2000668) |
| 2.4 | 11.8 | GO:0010958 | regulation of amino acid import(GO:0010958) L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 2.4 | 7.1 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 2.4 | 9.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 2.3 | 11.4 | GO:0070543 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 2.3 | 9.0 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 2.2 | 4.5 | GO:0090027 | negative regulation of monocyte chemotaxis(GO:0090027) |
| 2.1 | 10.6 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 2.0 | 6.1 | GO:0070104 | negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 2.0 | 5.9 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 1.9 | 5.7 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 1.8 | 7.4 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 1.8 | 43.0 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 1.8 | 12.4 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 1.8 | 10.6 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 1.7 | 79.5 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 1.7 | 3.3 | GO:1990051 | activation of protein kinase C activity(GO:1990051) |
| 1.5 | 3.1 | GO:0002859 | negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) |
| 1.5 | 4.6 | GO:0044206 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) CMP metabolic process(GO:0046035) |
| 1.5 | 36.1 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 1.5 | 25.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 1.4 | 7.2 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 1.4 | 4.2 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 1.4 | 13.9 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 1.4 | 4.2 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 1.3 | 5.3 | GO:0046226 | coumarin catabolic process(GO:0046226) |
| 1.3 | 10.5 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 1.3 | 1.3 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 1.3 | 2.6 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 1.3 | 6.5 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) |
| 1.3 | 42.1 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 1.3 | 3.8 | GO:0035874 | amiloride transport(GO:0015898) cellular response to copper ion starvation(GO:0035874) response to azide(GO:0097184) cellular response to azide(GO:0097185) |
| 1.3 | 6.3 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 1.3 | 12.6 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 1.3 | 41.5 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 1.2 | 9.9 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 1.2 | 3.7 | GO:0021558 | trochlear nerve development(GO:0021558) auditory receptor cell fate determination(GO:0042668) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 1.1 | 11.4 | GO:0002669 | positive regulation of T cell tolerance induction(GO:0002666) positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 1.1 | 5.5 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 1.0 | 8.3 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 1.0 | 7.0 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 1.0 | 9.9 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 1.0 | 7.7 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.9 | 7.3 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.9 | 11.7 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.9 | 29.5 | GO:0048535 | lymph node development(GO:0048535) |
| 0.9 | 7.1 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.9 | 2.7 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.9 | 4.4 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.9 | 4.4 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.8 | 6.8 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.8 | 4.2 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.8 | 15.9 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.8 | 14.2 | GO:0043320 | natural killer cell degranulation(GO:0043320) |
| 0.8 | 9.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.8 | 3.3 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.8 | 4.1 | GO:0035546 | detection of virus(GO:0009597) interferon-beta secretion(GO:0035546) regulation of interferon-beta secretion(GO:0035547) positive regulation of interferon-beta secretion(GO:0035549) |
| 0.8 | 2.4 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.8 | 20.2 | GO:0038065 | collagen-activated signaling pathway(GO:0038065) |
| 0.8 | 6.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.8 | 15.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.8 | 2.3 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.8 | 15.9 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.7 | 2.2 | GO:0072107 | regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) |
| 0.7 | 7.2 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.7 | 2.1 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.7 | 2.1 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.7 | 6.4 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.7 | 9.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.7 | 6.2 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.7 | 2.0 | GO:1901738 | regulation of vitamin A metabolic process(GO:1901738) |
| 0.7 | 2.7 | GO:0009223 | dUMP biosynthetic process(GO:0006226) pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.7 | 2.0 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.7 | 2.6 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.6 | 1.9 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.6 | 2.5 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.6 | 1.9 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.6 | 53.4 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.6 | 20.8 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.6 | 1.8 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.6 | 6.0 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.6 | 14.7 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.6 | 3.5 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.6 | 7.0 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.6 | 6.3 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.6 | 2.8 | GO:0070859 | lipid transport involved in lipid storage(GO:0010877) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.6 | 2.8 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.5 | 3.2 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.5 | 3.2 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.5 | 3.7 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.5 | 2.1 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.5 | 2.1 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.5 | 2.5 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.5 | 2.5 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.5 | 1.0 | GO:0002730 | regulation of dendritic cell cytokine production(GO:0002730) |
| 0.5 | 2.9 | GO:2000121 | regulation of removal of superoxide radicals(GO:2000121) |
| 0.5 | 1.5 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.5 | 3.3 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.5 | 5.2 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.5 | 2.8 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.5 | 2.8 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.5 | 1.4 | GO:0042374 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.5 | 0.9 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.4 | 1.3 | GO:1903699 | tarsal gland development(GO:1903699) |
| 0.4 | 4.0 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.4 | 5.3 | GO:0071236 | cellular response to antibiotic(GO:0071236) |
| 0.4 | 1.3 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.4 | 16.0 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.4 | 3.8 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.4 | 10.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.4 | 2.1 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.4 | 2.9 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.4 | 2.1 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.4 | 4.0 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.4 | 3.6 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.4 | 2.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.4 | 16.4 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.4 | 1.6 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.4 | 1.5 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.4 | 3.8 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.4 | 1.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.4 | 7.8 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.4 | 2.9 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.4 | 8.6 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.4 | 2.1 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.3 | 2.8 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.3 | 4.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.3 | 2.0 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.3 | 2.7 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.3 | 1.3 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.3 | 9.3 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.3 | 6.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.3 | 1.7 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.3 | 3.9 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.3 | 1.6 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.3 | 3.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.3 | 19.9 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.3 | 2.2 | GO:0033034 | positive regulation of neutrophil apoptotic process(GO:0033031) positive regulation of myeloid cell apoptotic process(GO:0033034) |
| 0.3 | 5.1 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.3 | 1.6 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.3 | 2.4 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.3 | 1.1 | GO:0015744 | succinate transport(GO:0015744) |
| 0.3 | 3.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.3 | 1.8 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.3 | 1.8 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.3 | 1.0 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.2 | 1.2 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.2 | 1.5 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.2 | 2.9 | GO:0097284 | hepatocyte apoptotic process(GO:0097284) |
| 0.2 | 0.7 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.2 | 0.7 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.2 | 2.7 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 | 1.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.2 | 0.7 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.2 | 1.2 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.2 | 0.9 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.2 | 3.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 0.7 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.2 | 3.4 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.2 | 2.2 | GO:0008228 | opsonization(GO:0008228) |
| 0.2 | 11.5 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.2 | 1.3 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.2 | 33.1 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.2 | 1.7 | GO:0032735 | positive regulation of interleukin-12 production(GO:0032735) |
| 0.2 | 17.4 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.2 | 3.0 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.2 | 3.4 | GO:0070633 | transepithelial transport(GO:0070633) |
| 0.2 | 1.9 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.2 | 0.8 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.2 | 1.7 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.2 | 2.8 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.2 | 1.4 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.2 | 1.4 | GO:0060023 | soft palate development(GO:0060023) |
| 0.2 | 1.2 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.2 | 1.0 | GO:0060931 | sinoatrial node cell differentiation(GO:0060921) sinoatrial node cell development(GO:0060931) |
| 0.2 | 2.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.2 | 4.5 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.2 | 0.2 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.2 | 4.7 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.2 | 1.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 0.7 | GO:0030091 | protein repair(GO:0030091) |
| 0.2 | 2.8 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.2 | 0.5 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.2 | 2.1 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.2 | 1.2 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.2 | 1.0 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.2 | 0.7 | GO:0001562 | response to protozoan(GO:0001562) |
| 0.2 | 5.5 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.2 | 1.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 0.9 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.2 | 2.5 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.2 | 3.3 | GO:0051923 | sulfation(GO:0051923) |
| 0.2 | 2.2 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.1 | 8.7 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 0.8 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.1 | 1.4 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 2.0 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 1.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 5.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 1.6 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.1 | 1.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.9 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 7.3 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.1 | 4.9 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 1.3 | GO:0098706 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.1 | 3.5 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 2.5 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.1 | 1.7 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.1 | 1.9 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) |
| 0.1 | 1.1 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.1 | 2.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 1.6 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.7 | GO:0006901 | vesicle coating(GO:0006901) |
| 0.1 | 2.2 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 1.1 | GO:0036006 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.1 | 8.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 2.9 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.1 | 10.3 | GO:0042098 | T cell proliferation(GO:0042098) |
| 0.1 | 0.5 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.1 | 3.1 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.1 | 0.3 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 0.1 | 0.7 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 2.3 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.1 | 8.0 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 0.5 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 8.9 | GO:0050868 | negative regulation of T cell activation(GO:0050868) |
| 0.1 | 15.3 | GO:0009615 | response to virus(GO:0009615) |
| 0.1 | 1.4 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 8.3 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.1 | 1.0 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.1 | 5.1 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.1 | 2.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 1.1 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 3.3 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 0.6 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 1.0 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 3.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.1 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 0.3 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 1.4 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 2.0 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.1 | GO:2000229 | pancreatic stellate cell proliferation(GO:0072343) regulation of pancreatic stellate cell proliferation(GO:2000229) |
| 0.1 | 3.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 1.8 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 0.9 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 | 3.9 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 1.7 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 5.2 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 10.4 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.1 | 1.6 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.1 | 1.8 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.1 | 0.5 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.1 | 1.3 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.1 | 0.7 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 4.0 | GO:0043297 | apical junction assembly(GO:0043297) |
| 0.1 | 0.7 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.1 | 0.8 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.1 | 1.5 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.6 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.7 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 0.2 | GO:0001916 | positive regulation of T cell mediated cytotoxicity(GO:0001916) |
| 0.1 | 0.8 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 10.7 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 2.1 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.1 | 0.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 2.6 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 0.9 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 4.9 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 0.3 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.1 | 1.8 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 2.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 2.5 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 | 0.3 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 6.0 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 2.6 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.5 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 7.8 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.0 | 0.2 | GO:0060318 | definitive erythrocyte differentiation(GO:0060318) |
| 0.0 | 1.0 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.9 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 0.6 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 2.9 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.4 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 0.2 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.3 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 1.8 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 1.1 | GO:0048701 | embryonic cranial skeleton morphogenesis(GO:0048701) |
| 0.0 | 0.4 | GO:0072431 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) |
| 0.0 | 0.9 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.0 | 0.1 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.8 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.0 | 1.1 | GO:0035088 | establishment or maintenance of apical/basal cell polarity(GO:0035088) establishment or maintenance of bipolar cell polarity(GO:0061245) |
| 0.0 | 3.5 | GO:2001236 | regulation of extrinsic apoptotic signaling pathway(GO:2001236) |
| 0.0 | 1.6 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.5 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 1.2 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 1.0 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 4.8 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 0.8 | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle(GO:0007091) metaphase/anaphase transition of cell cycle(GO:0044784) |
| 0.0 | 0.8 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.4 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.5 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 | 0.2 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.8 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.2 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.1 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.4 | GO:0007032 | endosome organization(GO:0007032) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.1 | 70.4 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 6.6 | 26.5 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 5.8 | 81.8 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 5.8 | 69.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 5.6 | 45.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 4.7 | 28.0 | GO:0031523 | Myb complex(GO:0031523) |
| 4.6 | 13.7 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 3.5 | 10.6 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 3.3 | 3.3 | GO:0042611 | MHC protein complex(GO:0042611) |
| 3.1 | 12.4 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 2.4 | 14.5 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 2.3 | 20.9 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 2.3 | 9.2 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 2.0 | 26.1 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 1.8 | 14.7 | GO:0042825 | TAP complex(GO:0042825) |
| 1.3 | 5.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 1.0 | 7.3 | GO:0098536 | deuterosome(GO:0098536) |
| 1.0 | 5.1 | GO:0031251 | PAN complex(GO:0031251) |
| 1.0 | 25.6 | GO:0000145 | exocyst(GO:0000145) |
| 1.0 | 16.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 1.0 | 10.5 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.9 | 4.5 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.9 | 6.3 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.8 | 9.8 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.8 | 2.4 | GO:1904602 | serotonin-activated cation-selective channel complex(GO:1904602) |
| 0.8 | 16.2 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.7 | 13.1 | GO:0036038 | MKS complex(GO:0036038) |
| 0.7 | 12.2 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.7 | 2.1 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.7 | 11.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.7 | 7.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.7 | 13.9 | GO:0042581 | specific granule(GO:0042581) |
| 0.7 | 4.6 | GO:0098642 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.7 | 2.0 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.6 | 2.9 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.6 | 24.6 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.5 | 32.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.4 | 2.1 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.4 | 1.5 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.4 | 3.4 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.3 | 1.0 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.3 | 3.2 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.3 | 1.6 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.3 | 178.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.3 | 1.2 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.3 | 1.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.3 | 2.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.3 | 3.3 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.3 | 14.2 | GO:0002102 | podosome(GO:0002102) |
| 0.3 | 6.3 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.3 | 7.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.3 | 2.8 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.2 | 0.7 | GO:1904511 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.2 | 2.2 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.2 | 1.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 2.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.2 | 1.8 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.2 | 2.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 18.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 1.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.2 | 1.8 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.2 | 2.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 2.9 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.2 | 2.5 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.2 | 9.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 8.0 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.2 | 32.6 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.2 | 0.7 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.2 | 3.0 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.2 | 11.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 9.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 4.1 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.2 | 2.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.2 | 2.5 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 20.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.2 | 8.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 1.6 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 7.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 28.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 0.5 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 2.2 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 15.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 2.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.4 | GO:1990427 | stereocilia tip link(GO:0002140) myosin VII complex(GO:0031477) stereocilia tip-link density(GO:1990427) upper tip-link density(GO:1990435) |
| 0.1 | 4.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 15.1 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 0.6 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.1 | 3.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.6 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 5.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 7.5 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 32.8 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 38.5 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 3.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 4.6 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 5.4 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 2.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 4.7 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.1 | 0.9 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 2.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 0.8 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 2.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 8.4 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.1 | 1.8 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 9.9 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 33.9 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.1 | 7.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 6.6 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 1.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.3 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 0.9 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 1.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 2.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 2.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 2.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 64.6 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 2.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.0 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 2.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 53.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 84.9 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 0.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 2.0 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 6.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.7 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0034451 | centriolar satellite(GO:0034451) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.2 | 42.6 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 8.2 | 24.7 | GO:0031726 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) CCR1 chemokine receptor binding(GO:0031726) |
| 8.2 | 48.9 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 6.6 | 26.5 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 5.4 | 96.5 | GO:0046977 | TAP binding(GO:0046977) |
| 5.1 | 30.6 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 4.7 | 19.0 | GO:0070976 | TIR domain binding(GO:0070976) |
| 4.7 | 28.0 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 4.6 | 13.7 | GO:0042497 | triacyl lipopeptide binding(GO:0042497) |
| 3.8 | 30.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 3.4 | 13.4 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 2.9 | 11.7 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 2.7 | 24.7 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 2.6 | 10.2 | GO:0019862 | IgA binding(GO:0019862) |
| 2.4 | 28.9 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 2.3 | 11.4 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 2.3 | 9.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 1.9 | 15.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 1.8 | 7.0 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 1.7 | 11.7 | GO:1990254 | keratin filament binding(GO:1990254) |
| 1.5 | 6.0 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 1.4 | 36.0 | GO:0070628 | proteasome binding(GO:0070628) |
| 1.4 | 4.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 1.3 | 5.3 | GO:0004063 | aryldialkylphosphatase activity(GO:0004063) |
| 1.3 | 3.8 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 1.3 | 77.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 1.3 | 3.8 | GO:0052597 | diamine oxidase activity(GO:0052597) histamine oxidase activity(GO:0052598) methylputrescine oxidase activity(GO:0052599) propane-1,3-diamine oxidase activity(GO:0052600) |
| 1.3 | 59.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 1.2 | 3.7 | GO:0071820 | N-box binding(GO:0071820) |
| 1.2 | 30.9 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 1.2 | 10.6 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 1.1 | 4.5 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 1.1 | 16.6 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 1.1 | 6.5 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 1.1 | 11.8 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 1.0 | 18.9 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.9 | 1.8 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.9 | 8.2 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.9 | 9.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.9 | 43.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.9 | 33.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.8 | 7.5 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.8 | 7.4 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.8 | 5.7 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.8 | 6.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.8 | 4.0 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.8 | 12.6 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.8 | 11.0 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.8 | 2.3 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.8 | 2.3 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.7 | 2.2 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.7 | 2.1 | GO:0061749 | forked DNA-dependent helicase activity(GO:0061749) flap-structured DNA binding(GO:0070336) |
| 0.7 | 7.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.7 | 11.6 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.7 | 3.4 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.7 | 2.6 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.7 | 4.6 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.7 | 2.6 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.6 | 7.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.6 | 8.6 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.6 | 2.4 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.6 | 2.4 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.6 | 2.9 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.6 | 1.8 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.6 | 5.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.6 | 4.0 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.5 | 2.2 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.5 | 9.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.5 | 14.2 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.5 | 3.9 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.5 | 2.4 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.5 | 2.9 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.5 | 3.3 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.5 | 2.7 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.4 | 14.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.4 | 2.6 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.4 | 9.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.4 | 32.3 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.4 | 2.5 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.4 | 1.7 | GO:0051435 | BH4 domain binding(GO:0051435) |
| 0.4 | 8.6 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.4 | 22.5 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.4 | 4.0 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.4 | 2.3 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.4 | 11.5 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.4 | 5.7 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.4 | 4.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.4 | 2.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.4 | 2.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.3 | 44.6 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.3 | 2.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.3 | 4.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.3 | 1.7 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.3 | 0.9 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.3 | 1.2 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.3 | 4.6 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.3 | 2.8 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.3 | 0.9 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.3 | 0.9 | GO:0045030 | UTP-activated nucleotide receptor activity(GO:0045030) |
| 0.3 | 4.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.3 | 4.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.3 | 9.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.3 | 2.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.3 | 10.3 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.3 | 1.2 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.3 | 1.4 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.3 | 3.3 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.3 | 2.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.3 | 1.9 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.3 | 1.3 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.3 | 1.9 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.3 | 2.4 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.3 | 1.1 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.3 | 1.8 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.3 | 6.5 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.3 | 3.3 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.3 | 8.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.2 | 1.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.2 | 1.7 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.2 | 4.9 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.2 | 0.7 | GO:0008396 | oxysterol 7-alpha-hydroxylase activity(GO:0008396) |
| 0.2 | 12.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 4.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 0.7 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.2 | 1.4 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.2 | 2.7 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.2 | 9.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.2 | 5.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 1.3 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.2 | 1.3 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.2 | 1.3 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.2 | 4.0 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.2 | 3.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.2 | 3.5 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.2 | 25.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 0.2 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.2 | 1.2 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.2 | 1.0 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.2 | 10.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 86.2 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.2 | 1.4 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.2 | 1.4 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.2 | 1.3 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 1.4 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.2 | 2.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 8.5 | GO:0003823 | antigen binding(GO:0003823) |
| 0.2 | 1.3 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 2.0 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.2 | 23.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.2 | 11.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.2 | 1.7 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 1.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.7 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 1.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 18.8 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.1 | 47.2 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 29.8 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) |
| 0.1 | 25.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 1.0 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 2.5 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 2.8 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 2.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 1.6 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 1.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 3.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.7 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 1.0 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.6 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 3.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 47.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 2.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 4.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.8 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.1 | 9.7 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.1 | 1.5 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.1 | 1.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 7.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 36.9 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 4.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 0.3 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 0.1 | 0.7 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.5 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 2.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 1.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 3.4 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.1 | 1.3 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.5 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 4.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 1.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 2.8 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 1.4 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 5.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 0.2 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.1 | 1.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 1.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 3.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 1.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.6 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 0.9 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 2.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.2 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.0 | 0.7 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 2.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 6.9 | GO:0005506 | iron ion binding(GO:0005506) |
| 0.0 | 1.2 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 3.7 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 1.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 6.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 5.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 2.8 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 1.9 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 1.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 1.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.4 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 8.0 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 31.5 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 0.4 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 1.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.7 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 211.8 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 1.8 | 45.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 1.7 | 14.0 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 1.6 | 61.6 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 1.4 | 18.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 1.1 | 50.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 1.0 | 14.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 1.0 | 26.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.8 | 36.2 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.8 | 15.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.7 | 8.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.6 | 34.4 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.6 | 20.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.6 | 33.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.5 | 32.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.5 | 24.3 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.5 | 34.8 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.5 | 3.7 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.5 | 26.6 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.4 | 29.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.4 | 10.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.4 | 11.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.3 | 6.8 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.3 | 2.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.3 | 5.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.3 | 12.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.2 | 10.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.2 | 38.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.2 | 11.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 4.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.2 | 14.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 8.9 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.2 | 6.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.2 | 2.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.2 | 4.8 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.2 | 4.4 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.2 | 11.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.2 | 3.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.2 | 1.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 2.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 4.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 25.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 10.0 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 2.4 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 1.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 1.8 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 2.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 0.6 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 2.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 2.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 2.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 1.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 1.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 4.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 2.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 2.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 4.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 2.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 4.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.3 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.4 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.2 | PID ARF6 PATHWAY | Arf6 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.8 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 3.4 | 13.7 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 3.4 | 87.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 3.0 | 121.7 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 2.2 | 118.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 1.8 | 42.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.9 | 47.6 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.9 | 11.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.9 | 9.8 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.7 | 21.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.6 | 17.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.6 | 3.0 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.6 | 8.2 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.5 | 5.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.5 | 10.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.5 | 9.0 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.5 | 4.0 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.5 | 3.8 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.5 | 12.0 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.4 | 3.1 | REACTOME P75NTR SIGNALS VIA NFKB | Genes involved in p75NTR signals via NF-kB |
| 0.4 | 27.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.4 | 38.1 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.4 | 17.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.4 | 12.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.4 | 13.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.4 | 10.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.3 | 36.0 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.3 | 1.5 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.3 | 11.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.3 | 11.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 29.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.2 | 8.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 6.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 6.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 2.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 3.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 7.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.2 | 8.9 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.2 | 3.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.2 | 7.7 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.2 | 3.3 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.2 | 2.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 2.1 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 5.2 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 3.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 2.9 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 5.7 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 4.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 3.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 2.4 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 5.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 11.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 10.9 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.1 | 1.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 7.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 8.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 2.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 10.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 5.8 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.1 | 0.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 2.5 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 2.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 3.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 3.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 2.2 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.1 | 0.9 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 5.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.0 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.5 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.9 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 2.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.6 | REACTOME MEMBRANE TRAFFICKING | Genes involved in Membrane Trafficking |
| 0.0 | 0.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |