Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Relb
Z-value: 1.00
Transcription factors associated with Relb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Relb
|
ENSMUSG00000002983.18 | Relb |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Relb | mm39_v1_chr7_-_19363280_19363372 | 0.56 | 3.7e-07 | Click! |
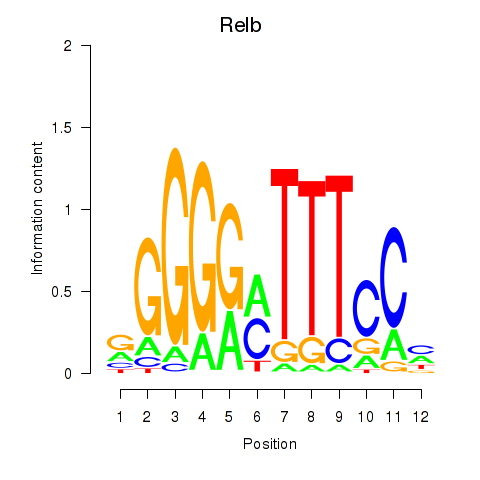
Activity profile of Relb motif
Sorted Z-values of Relb motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Relb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_37504783 | 11.73 |
ENSMUST00000038844.7
|
Ubd
|
ubiquitin D |
| chr6_+_68233361 | 11.58 |
ENSMUST00000103320.3
|
Igkv14-111
|
immunoglobulin kappa variable 14-111 |
| chr6_-_69415741 | 8.49 |
ENSMUST00000103354.3
|
Igkv4-59
|
immunoglobulin kappa variable 4-59 |
| chr17_+_35413415 | 8.24 |
ENSMUST00000025262.6
ENSMUST00000173600.2 |
Ltb
|
lymphotoxin B |
| chr1_+_135060431 | 7.87 |
ENSMUST00000187985.7
ENSMUST00000049449.11 |
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr6_-_68812291 | 6.91 |
ENSMUST00000199143.5
ENSMUST00000103335.3 |
Igkv12-89
|
immunoglobulin kappa chain variable 12-89 |
| chr6_-_69792108 | 6.60 |
ENSMUST00000103367.3
|
Igkv12-44
|
immunoglobulin kappa variable 12-44 |
| chr1_+_135060994 | 6.16 |
ENSMUST00000167080.3
|
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chrX_+_156482116 | 6.13 |
ENSMUST00000112521.8
|
Smpx
|
small muscle protein, X-linked |
| chr6_+_67873135 | 6.06 |
ENSMUST00000103310.3
|
Igkv14-126
|
immunoglobulin kappa variable 14-126 |
| chrX_+_156481906 | 5.77 |
ENSMUST00000136141.2
ENSMUST00000190091.7 |
Smpx
|
small muscle protein, X-linked |
| chr6_-_69835868 | 5.25 |
ENSMUST00000103369.2
|
Igkv12-41
|
immunoglobulin kappa chain variable 12-41 |
| chr2_+_13579092 | 5.23 |
ENSMUST00000193675.2
|
Vim
|
vimentin |
| chr17_+_31427023 | 5.22 |
ENSMUST00000173776.2
ENSMUST00000048656.15 |
Ubash3a
|
ubiquitin associated and SH3 domain containing, A |
| chr10_-_18891095 | 4.84 |
ENSMUST00000019997.11
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr10_+_96452860 | 4.67 |
ENSMUST00000038377.9
|
Btg1
|
BTG anti-proliferation factor 1 |
| chr2_+_13578738 | 4.66 |
ENSMUST00000141365.3
ENSMUST00000028062.8 |
Vim
|
vimentin |
| chr7_-_19363280 | 4.00 |
ENSMUST00000094762.10
ENSMUST00000049912.15 ENSMUST00000098754.5 |
Relb
|
avian reticuloendotheliosis viral (v-rel) oncogene related B |
| chr2_-_34848148 | 3.98 |
ENSMUST00000201690.2
ENSMUST00000172159.8 |
Traf1
|
TNF receptor-associated factor 1 |
| chr2_-_76812799 | 3.95 |
ENSMUST00000011934.13
ENSMUST00000099981.10 ENSMUST00000099980.10 ENSMUST00000111882.9 ENSMUST00000140091.8 |
Ttn
|
titin |
| chr10_+_79500387 | 3.28 |
ENSMUST00000020554.8
|
Madcam1
|
mucosal vascular addressin cell adhesion molecule 1 |
| chr12_+_76417040 | 3.24 |
ENSMUST00000042779.4
|
Zbtb1
|
zinc finger and BTB domain containing 1 |
| chr19_+_46293210 | 3.21 |
ENSMUST00000236591.2
|
Nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 |
| chr7_-_45570254 | 3.18 |
ENSMUST00000164119.3
|
Emp3
|
epithelial membrane protein 3 |
| chr4_-_148584906 | 3.17 |
ENSMUST00000030840.4
|
Angptl7
|
angiopoietin-like 7 |
| chr11_-_23720953 | 3.14 |
ENSMUST00000102864.5
|
Rel
|
reticuloendotheliosis oncogene |
| chr10_+_79500421 | 3.14 |
ENSMUST00000217748.2
|
Madcam1
|
mucosal vascular addressin cell adhesion molecule 1 |
| chr12_-_76416155 | 3.12 |
ENSMUST00000176187.8
ENSMUST00000167011.8 ENSMUST00000176967.2 |
Zbtb25
|
zinc finger and BTB domain containing 25 |
| chr6_+_68547717 | 2.99 |
ENSMUST00000196839.5
ENSMUST00000103327.3 |
Igkv12-98
|
immunoglobulin kappa variable 12-98 |
| chr2_-_5719302 | 2.96 |
ENSMUST00000044009.14
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr7_-_45570538 | 2.94 |
ENSMUST00000210297.2
|
Emp3
|
epithelial membrane protein 3 |
| chr12_-_76416247 | 2.84 |
ENSMUST00000176278.8
|
Zbtb25
|
zinc finger and BTB domain containing 25 |
| chr12_+_85733415 | 2.72 |
ENSMUST00000040536.6
|
Batf
|
basic leucine zipper transcription factor, ATF-like |
| chr17_+_29020064 | 2.58 |
ENSMUST00000004985.11
|
Brpf3
|
bromodomain and PHD finger containing, 3 |
| chr5_-_123127346 | 2.53 |
ENSMUST00000118027.8
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr19_+_46293160 | 2.44 |
ENSMUST00000073116.13
|
Nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 |
| chr13_+_44994167 | 2.43 |
ENSMUST00000173906.3
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr7_-_99276310 | 2.34 |
ENSMUST00000178124.3
|
Tpbgl
|
trophoblast glycoprotein-like |
| chr3_+_51323383 | 2.29 |
ENSMUST00000029303.13
|
Naa15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr6_+_67890534 | 2.22 |
ENSMUST00000197406.5
ENSMUST00000103311.3 |
Igkv11-125
|
immunoglobulin kappa variable 11-125 |
| chr12_+_111383864 | 2.18 |
ENSMUST00000220537.2
ENSMUST00000223050.2 ENSMUST00000072646.8 ENSMUST00000223431.2 ENSMUST00000221144.2 ENSMUST00000222437.2 |
Exoc3l4
|
exocyst complex component 3-like 4 |
| chr4_+_129713991 | 2.17 |
ENSMUST00000030578.14
|
Ptp4a2
|
protein tyrosine phosphatase 4a2 |
| chr4_-_152216322 | 2.13 |
ENSMUST00000105653.8
|
Espn
|
espin |
| chr11_+_69471219 | 2.10 |
ENSMUST00000108657.4
|
Trp53
|
transformation related protein 53 |
| chr4_+_151012375 | 2.03 |
ENSMUST00000139826.8
ENSMUST00000116257.8 |
Tnfrsf9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr11_+_69471185 | 1.96 |
ENSMUST00000171247.8
ENSMUST00000108658.10 ENSMUST00000005371.12 |
Trp53
|
transformation related protein 53 |
| chr2_-_73316053 | 1.91 |
ENSMUST00000102680.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr8_+_72128920 | 1.88 |
ENSMUST00000110013.10
ENSMUST00000051995.14 |
Jak3
|
Janus kinase 3 |
| chr8_+_47192911 | 1.70 |
ENSMUST00000208433.2
|
Irf2
|
interferon regulatory factor 2 |
| chr10_-_93146825 | 1.63 |
ENSMUST00000151153.2
|
Elk3
|
ELK3, member of ETS oncogene family |
| chr12_-_55539372 | 1.58 |
ENSMUST00000021413.9
|
Nfkbia
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, alpha |
| chr11_-_84761009 | 1.57 |
ENSMUST00000154915.9
|
Ggnbp2
|
gametogenetin binding protein 2 |
| chr2_+_18677195 | 1.52 |
ENSMUST00000171845.8
ENSMUST00000061158.5 |
Commd3
|
COMM domain containing 3 |
| chr11_+_100751151 | 1.50 |
ENSMUST00000138083.8
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chr16_+_91343936 | 1.50 |
ENSMUST00000023687.9
|
Ifngr2
|
interferon gamma receptor 2 |
| chr2_-_160155536 | 1.47 |
ENSMUST00000109475.3
|
Gm826
|
predicted gene 826 |
| chr10_-_93146937 | 1.47 |
ENSMUST00000008542.12
|
Elk3
|
ELK3, member of ETS oncogene family |
| chr4_-_116982804 | 1.42 |
ENSMUST00000183310.2
|
Btbd19
|
BTB (POZ) domain containing 19 |
| chr11_-_84761472 | 1.38 |
ENSMUST00000018547.9
|
Ggnbp2
|
gametogenetin binding protein 2 |
| chr6_-_69920632 | 1.33 |
ENSMUST00000198880.5
ENSMUST00000103371.3 |
Igkv12-38
|
immunoglobulin kappa chain variable 12-38 |
| chr11_-_54846873 | 1.30 |
ENSMUST00000155316.2
ENSMUST00000108889.10 ENSMUST00000126703.8 |
Tnip1
|
TNFAIP3 interacting protein 1 |
| chr5_+_90907207 | 1.28 |
ENSMUST00000031318.6
|
Cxcl5
|
chemokine (C-X-C motif) ligand 5 |
| chr10_-_79940168 | 1.28 |
ENSMUST00000219260.2
|
Sbno2
|
strawberry notch 2 |
| chr11_-_84761538 | 1.18 |
ENSMUST00000170741.2
ENSMUST00000172405.8 ENSMUST00000100686.10 ENSMUST00000108081.9 |
Ggnbp2
|
gametogenetin binding protein 2 |
| chr8_-_21392510 | 1.15 |
ENSMUST00000122025.9
|
Gm15056
|
predicted gene 15056 |
| chr17_-_71153283 | 1.12 |
ENSMUST00000156484.2
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr12_-_84240781 | 1.12 |
ENSMUST00000110294.2
|
Mideas
|
mitotic deacetylase associated SANT domain protein |
| chr1_+_132996237 | 1.12 |
ENSMUST00000239467.2
|
Pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
| chr2_-_118377500 | 1.12 |
ENSMUST00000125860.3
|
Bmf
|
BCL2 modifying factor |
| chr8_+_47192767 | 1.03 |
ENSMUST00000034041.9
ENSMUST00000208507.2 ENSMUST00000207105.2 |
Irf2
|
interferon regulatory factor 2 |
| chr14_+_53878403 | 1.03 |
ENSMUST00000184874.2
|
Trav14-2
|
T cell receptor alpha variable 14-2 |
| chr5_-_123127148 | 0.97 |
ENSMUST00000046073.16
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr2_-_5068807 | 0.96 |
ENSMUST00000114996.8
|
Optn
|
optineurin |
| chr11_-_84761637 | 0.88 |
ENSMUST00000168434.8
|
Ggnbp2
|
gametogenetin binding protein 2 |
| chr17_+_34129221 | 0.84 |
ENSMUST00000174541.9
|
Daxx
|
Fas death domain-associated protein |
| chr2_-_5068743 | 0.83 |
ENSMUST00000027986.5
|
Optn
|
optineurin |
| chr14_-_55828511 | 0.74 |
ENSMUST00000161807.8
ENSMUST00000111378.10 ENSMUST00000159687.2 |
Psme2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chrX_+_162692126 | 0.66 |
ENSMUST00000033734.14
ENSMUST00000112294.9 |
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chrX_-_133012457 | 0.50 |
ENSMUST00000159259.3
ENSMUST00000113275.10 |
Nox1
|
NADPH oxidase 1 |
| chr2_+_119803230 | 0.49 |
ENSMUST00000229024.2
|
Mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr16_+_17712061 | 0.48 |
ENSMUST00000046937.4
|
Tssk1
|
testis-specific serine kinase 1 |
| chr17_+_29020744 | 0.45 |
ENSMUST00000136233.2
|
Brpf3
|
bromodomain and PHD finger containing, 3 |
| chr12_+_41074328 | 0.44 |
ENSMUST00000134965.8
|
Immp2l
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr2_-_37312881 | 0.35 |
ENSMUST00000112936.4
ENSMUST00000112934.8 |
Rc3h2
|
ring finger and CCCH-type zinc finger domains 2 |
| chr2_+_119803180 | 0.32 |
ENSMUST00000066058.8
|
Mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr13_+_83672389 | 0.31 |
ENSMUST00000200394.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr10_+_23952398 | 0.24 |
ENSMUST00000051133.6
|
Taar8a
|
trace amine-associated receptor 8A |
| chr12_+_41074089 | 0.22 |
ENSMUST00000132121.8
|
Immp2l
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr9_-_77452109 | 0.20 |
ENSMUST00000183734.2
|
Lrrc1
|
leucine rich repeat containing 1 |
| chr5_-_37981919 | 0.20 |
ENSMUST00000063116.10
|
Msx1
|
msh homeobox 1 |
| chr11_+_102775991 | 0.15 |
ENSMUST00000100369.4
|
Fam187a
|
family with sequence similarity 187, member A |
| chr12_+_111132908 | 0.13 |
ENSMUST00000139162.8
ENSMUST00000060274.7 |
Traf3
|
TNF receptor-associated factor 3 |
| chr5_+_91039092 | 0.08 |
ENSMUST00000031327.9
|
Cxcl1
|
chemokine (C-X-C motif) ligand 1 |
| chr9_-_77452152 | 0.06 |
ENSMUST00000183873.8
|
Lrrc1
|
leucine rich repeat containing 1 |
| chr18_-_43526411 | 0.05 |
ENSMUST00000025379.14
|
Dpysl3
|
dihydropyrimidinase-like 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0070429 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 1.4 | 4.1 | GO:1904024 | negative regulation of NAD metabolic process(GO:1902689) negative regulation of glucose catabolic process to lactate via pyruvate(GO:1904024) |
| 1.3 | 11.4 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 1.2 | 9.9 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 1.1 | 5.3 | GO:0002266 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.9 | 11.7 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.9 | 3.5 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.8 | 5.0 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.8 | 3.2 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.8 | 4.0 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.6 | 1.9 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.5 | 1.6 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.5 | 4.7 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.5 | 1.4 | GO:0070949 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.4 | 1.5 | GO:0060376 | positive regulation of mast cell differentiation(GO:0060376) |
| 0.4 | 1.1 | GO:1904092 | regulation of autophagic cell death(GO:1904092) negative regulation of autophagic cell death(GO:1904093) |
| 0.3 | 1.3 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.3 | 1.8 | GO:0061734 | negative regulation of receptor recycling(GO:0001920) parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.3 | 3.0 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.3 | 1.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.3 | 2.1 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.3 | 6.7 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.3 | 2.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.3 | 2.0 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.2 | 0.7 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.2 | 6.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.2 | 0.8 | GO:0072738 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.2 | 1.3 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.2 | 2.4 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.2 | 6.4 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.2 | 5.2 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 2.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.8 | GO:2000483 | activation of JNKK activity(GO:0007256) negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.1 | 0.3 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.1 | 21.9 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 0.2 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.1 | 1.9 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 1.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 16.5 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 3.0 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.1 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 7.3 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.0 | 4.0 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.7 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 | 2.8 | GO:0051607 | defense response to virus(GO:0051607) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 11.9 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 1.1 | 5.3 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.9 | 9.9 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.9 | 7.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.3 | 3.0 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.2 | 0.7 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.2 | 11.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.2 | 2.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 4.0 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 4.1 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 2.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 2.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 2.1 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 3.5 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 14.8 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 1.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.8 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.8 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 1.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 2.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 24.7 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.4 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 1.4 | 9.9 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.8 | 3.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.6 | 4.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.5 | 4.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.5 | 11.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.3 | 4.0 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.2 | 10.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 1.5 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.2 | 5.0 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.2 | 3.5 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 2.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 1.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.7 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 16.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 2.4 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 1.1 | GO:0001727 | lipid kinase activity(GO:0001727) 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 2.0 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 2.0 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 3.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 3.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 1.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 5.2 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 1.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 4.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 5.5 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 2.2 | GO:0000149 | SNARE binding(GO:0000149) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 14.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.3 | 6.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 11.3 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.2 | 9.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.9 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 4.8 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.1 | 3.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 5.5 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 2.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 2.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 4.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 9.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 6.5 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 3.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 4.8 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 2.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 1.5 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 3.9 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 1.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 1.1 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 3.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |