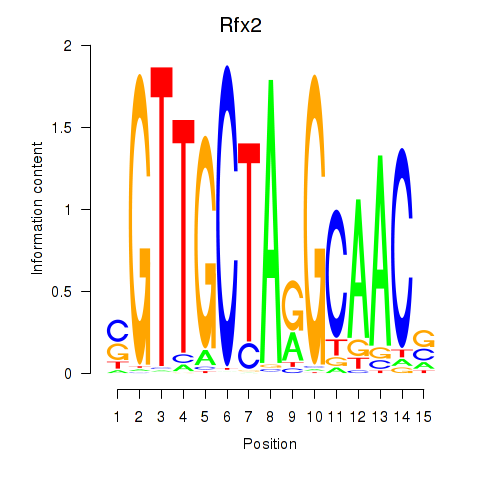
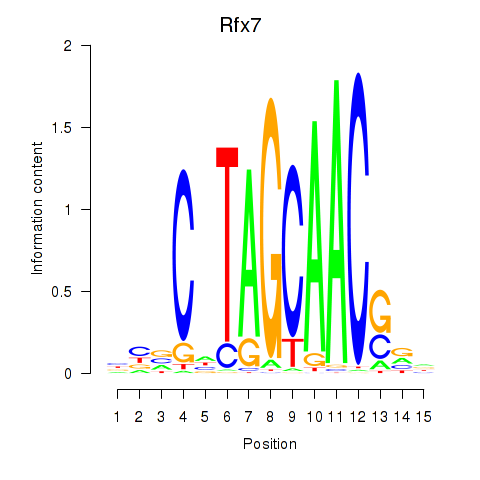
|
chr8_+_117231712
Show fit
|
52.96 |
ENSMUST00000213007.2
ENSMUST00000078170.7
|
Dynlrb2
|
dynein light chain roadblock-type 2
|
|
chr2_-_3420056
Show fit
|
50.55 |
ENSMUST00000115084.8
ENSMUST00000115083.8
|
Meig1
|
meiosis expressed gene 1
|
|
chr2_-_3420102
Show fit
|
50.12 |
ENSMUST00000115082.10
|
Meig1
|
meiosis expressed gene 1
|
|
chr12_+_33444853
Show fit
|
48.57 |
ENSMUST00000020878.8
|
Efcab10
|
EF-hand calcium binding domain 10
|
|
chr2_-_21210151
Show fit
|
46.54 |
ENSMUST00000027992.3
|
Enkur
|
enkurin, TRPC channel interacting protein
|
|
chr17_+_25690538
Show fit
|
45.64 |
ENSMUST00000234449.2
ENSMUST00000025002.3
ENSMUST00000235033.2
|
Tekt4
|
tektin 4
|
|
chr3_+_108444837
Show fit
|
45.53 |
ENSMUST00000029485.6
|
1700013F07Rik
|
RIKEN cDNA 1700013F07 gene
|
|
chr17_-_11059172
Show fit
|
44.90 |
ENSMUST00000041463.7
|
Pacrg
|
PARK2 co-regulated
|
|
chr11_-_96932220
Show fit
|
42.54 |
ENSMUST00000021251.7
|
Lrrc46
|
leucine rich repeat containing 46
|
|
chr12_-_110807330
Show fit
|
42.21 |
ENSMUST00000177224.2
ENSMUST00000084974.11
ENSMUST00000070565.15
|
Mok
|
MOK protein kinase
|
|
chr5_+_30623379
Show fit
|
39.86 |
ENSMUST00000127815.2
ENSMUST00000031078.10
ENSMUST00000114743.2
|
Fam166c
|
family with sequence similarity 166, member C
|
|
chr11_+_60244132
Show fit
|
38.97 |
ENSMUST00000070805.13
ENSMUST00000094140.9
ENSMUST00000108723.9
ENSMUST00000108722.5
|
Drc3
|
dynein regulatory complex subunit 3
|
|
chr7_-_126574959
Show fit
|
38.85 |
ENSMUST00000206296.2
ENSMUST00000205437.3
|
Cdiptos
|
CDIP transferase, opposite strand
|
|
chr19_-_10460238
Show fit
|
37.39 |
ENSMUST00000235392.2
ENSMUST00000237522.2
ENSMUST00000038842.5
|
Ppp1r32
|
protein phosphatase 1, regulatory subunit 32
|
|
chr13_+_68745542
Show fit
|
36.73 |
ENSMUST00000222604.2
|
1700001L19Rik
|
RIKEN cDNA 1700001L19 gene
|
|
chr11_+_100436433
Show fit
|
36.38 |
ENSMUST00000092684.12
ENSMUST00000006976.8
|
Odad4
|
outer dynein arm complex subunit 4
|
|
chr7_+_4785936
Show fit
|
36.15 |
ENSMUST00000013235.6
|
Tmem190
|
transmembrane protein 190
|
|
chr13_+_68745528
Show fit
|
35.43 |
ENSMUST00000022007.8
|
1700001L19Rik
|
RIKEN cDNA 1700001L19 gene
|
|
chr4_-_126219465
Show fit
|
35.36 |
ENSMUST00000102616.8
|
Tekt2
|
tektin 2
|
|
chr10_-_74868360
Show fit
|
35.32 |
ENSMUST00000159994.2
ENSMUST00000179546.8
ENSMUST00000160450.8
ENSMUST00000160072.8
ENSMUST00000009214.10
ENSMUST00000166088.8
|
Rsph14
|
radial spoke head homolog 14 (Chlamydomonas)
|
|
chr19_+_47926086
Show fit
|
34.86 |
ENSMUST00000238163.2
ENSMUST00000066308.9
|
Cfap58
|
cilia and flagella associated protein 58
|
|
chr11_-_30421792
Show fit
|
34.73 |
ENSMUST00000041763.14
|
4930505A04Rik
|
RIKEN cDNA 4930505A04 gene
|
|
chr12_+_105672227
Show fit
|
34.31 |
ENSMUST00000040876.7
|
Ak7
|
adenylate kinase 7
|
|
chr9_+_119766672
Show fit
|
34.06 |
ENSMUST00000035100.6
|
Ttc21a
|
tetratricopeptide repeat domain 21A
|
|
chr5_+_123627365
Show fit
|
32.93 |
ENSMUST00000196809.5
ENSMUST00000094327.10
ENSMUST00000121444.2
|
Lrrc43
|
leucine rich repeat containing 43
|
|
chr5_-_116427003
Show fit
|
31.80 |
ENSMUST00000086483.4
ENSMUST00000050178.13
|
Ccdc60
|
coiled-coil domain containing 60
|
|
chr8_+_39472981
Show fit
|
31.68 |
ENSMUST00000239508.1
ENSMUST00000239509.1
|
TUSC3
|
tumor suppressor candidate 3
|
|
chr6_-_24515036
Show fit
|
30.75 |
ENSMUST00000052277.5
|
Iqub
|
IQ motif and ubiquitin domain containing
|
|
chr2_+_70339157
Show fit
|
29.77 |
ENSMUST00000100041.9
|
Erich2
|
glutamate rich 2
|
|
chr18_+_57666852
Show fit
|
29.50 |
ENSMUST00000079738.10
ENSMUST00000135806.8
ENSMUST00000127130.9
|
Ccdc192
|
coiled-coil domain containing 192
|
|
chr9_-_21913896
Show fit
|
29.07 |
ENSMUST00000044926.6
|
Odad3
|
outer dynein arm docking complex subunit 3
|
|
chr2_-_181340226
Show fit
|
28.81 |
ENSMUST00000052416.4
|
Lkaaear1
|
LKAAEAR motif containing 1 (IKAAEAR murine motif)
|
|
chr4_-_117109074
Show fit
|
28.69 |
ENSMUST00000165128.9
|
Armh1
|
armadillo-like helical domain containing 1
|
|
chr1_-_10079325
Show fit
|
28.55 |
ENSMUST00000176398.8
ENSMUST00000027049.10
|
Ppp1r42
|
protein phosphatase 1, regulatory subunit 42
|
|
chr1_+_46105898
Show fit
|
28.14 |
ENSMUST00000069293.10
|
Dnah7b
|
dynein, axonemal, heavy chain 7B
|
|
chr11_+_68477812
Show fit
|
28.12 |
ENSMUST00000154294.8
ENSMUST00000063006.12
|
Ccdc42
|
coiled-coil domain containing 42
|
|
chr8_+_46387647
Show fit
|
28.09 |
ENSMUST00000095326.10
|
Ccdc110
|
coiled-coil domain containing 110
|
|
chr11_+_102772030
Show fit
|
27.82 |
ENSMUST00000021307.10
ENSMUST00000159834.2
|
Ccdc103
|
coiled-coil domain containing 103
|
|
chr2_+_19204076
Show fit
|
27.76 |
ENSMUST00000114640.9
ENSMUST00000049255.7
|
Armc3
|
armadillo repeat containing 3
|
|
chr1_+_46106006
Show fit
|
27.01 |
ENSMUST00000238212.2
|
Dnah7b
|
dynein, axonemal, heavy chain 7B
|
|
chr2_+_70339175
Show fit
|
26.95 |
ENSMUST00000134607.8
|
Erich2
|
glutamate rich 2
|
|
chr2_-_28356294
Show fit
|
26.73 |
ENSMUST00000127683.2
ENSMUST00000086370.11
|
1700007K13Rik
|
RIKEN cDNA 1700007K13 gene
|
|
chr9_-_21913833
Show fit
|
26.15 |
ENSMUST00000115336.10
|
Odad3
|
outer dynein arm docking complex subunit 3
|
|
chr1_-_16163506
Show fit
|
26.14 |
ENSMUST00000145070.8
ENSMUST00000151004.2
|
4930444P10Rik
|
RIKEN cDNA 4930444P10 gene
|
|
chr1_+_46464625
Show fit
|
26.00 |
ENSMUST00000189749.7
|
Dnah7c
|
dynein, axonemal, heavy chain 7C
|
|
chr11_+_114618209
Show fit
|
25.99 |
ENSMUST00000069325.14
|
Dnai2
|
dynein axonemal intermediate chain 2
|
|
chr5_+_30486375
Show fit
|
25.95 |
ENSMUST00000101448.5
|
Drc1
|
dynein regulatory complex subunit 1
|
|
chr15_+_98606067
Show fit
|
25.57 |
ENSMUST00000003444.6
ENSMUST00000229471.2
|
Ccdc65
|
coiled-coil domain containing 65
|
|
chr8_-_79975199
Show fit
|
25.42 |
ENSMUST00000034109.6
|
1700011L22Rik
|
RIKEN cDNA 1700011L22 gene
|
|
chr4_-_126219434
Show fit
|
25.15 |
ENSMUST00000131113.8
|
Tekt2
|
tektin 2
|
|
chr6_-_73198608
Show fit
|
24.77 |
ENSMUST00000064948.13
ENSMUST00000114040.8
|
Dnah6
|
dynein, axonemal, heavy chain 6
|
|
chrX_+_51880056
Show fit
|
24.58 |
ENSMUST00000101588.2
|
Ccdc160
|
coiled-coil domain containing 160
|
|
chr5_-_5714196
Show fit
|
23.84 |
ENSMUST00000196165.5
ENSMUST00000061008.10
ENSMUST00000135252.3
ENSMUST00000054865.13
|
Cfap69
|
cilia and flagella associated protein 69
|
|
chr5_-_21850579
Show fit
|
23.64 |
ENSMUST00000051358.11
|
Fbxl13
|
F-box and leucine-rich repeat protein 13
|
|
chr8_-_120904179
Show fit
|
23.63 |
ENSMUST00000048786.13
|
Cibar2
|
CBY1 interacting BAR domain containing 2
|
|
chr5_-_21850539
Show fit
|
23.40 |
ENSMUST00000115234.2
|
Fbxl13
|
F-box and leucine-rich repeat protein 13
|
|
chr3_+_99792699
Show fit
|
23.26 |
ENSMUST00000164539.2
|
Spag17
|
sperm associated antigen 17
|
|
chr2_-_32665342
Show fit
|
22.99 |
ENSMUST00000161089.8
ENSMUST00000066478.9
ENSMUST00000161950.8
ENSMUST00000091059.12
|
Ttc16
|
tetratricopeptide repeat domain 16
|
|
chr7_-_89552785
Show fit
|
22.75 |
ENSMUST00000041195.6
|
Ccdc81
|
coiled-coil domain containing 81
|
|
chr12_+_112772530
Show fit
|
22.45 |
ENSMUST00000037014.11
ENSMUST00000177808.3
|
Clba1
|
clathrin binding box of aftiphilin containing 1
|
|
chr9_+_72866067
Show fit
|
22.39 |
ENSMUST00000098567.9
ENSMUST00000034734.9
|
Dnaaf4
|
dynein axonemal assembly factor 4
|
|
chr4_-_62389098
Show fit
|
22.27 |
ENSMUST00000135811.2
ENSMUST00000120095.8
ENSMUST00000030087.14
ENSMUST00000107452.8
ENSMUST00000155522.8
|
Wdr31
|
WD repeat domain 31
|
|
chr7_-_139558599
Show fit
|
22.05 |
ENSMUST00000209335.2
ENSMUST00000210254.2
ENSMUST00000121412.2
ENSMUST00000097970.9
|
Spef1l
|
sperm flagellar 1 like
|
|
chr8_-_117705647
Show fit
|
22.04 |
ENSMUST00000070577.7
ENSMUST00000213068.2
|
1700030J22Rik
|
RIKEN cDNA 1700030J22 gene
|
|
chr3_-_145813802
Show fit
|
21.98 |
ENSMUST00000160285.2
|
Dnai3
|
dynein axonemal intermediate chain 3
|
|
chr1_-_131066004
Show fit
|
21.74 |
ENSMUST00000016670.9
|
Dyrk3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3
|
|
chr11_+_119119925
Show fit
|
21.42 |
ENSMUST00000053440.8
|
Ccdc40
|
coiled-coil domain containing 40
|
|
chr8_-_55177352
Show fit
|
21.37 |
ENSMUST00000129132.3
ENSMUST00000150488.8
ENSMUST00000127511.9
|
Wdr17
|
WD repeat domain 17
|
|
chr9_+_22322802
Show fit
|
21.13 |
ENSMUST00000058868.9
|
9530077C05Rik
|
RIKEN cDNA 9530077C05 gene
|
|
chr4_+_137989526
Show fit
|
20.79 |
ENSMUST00000030539.10
|
Kif17
|
kinesin family member 17
|
|
chr14_-_64225223
Show fit
|
20.37 |
ENSMUST00000022532.6
|
4930578I06Rik
|
RIKEN cDNA 4930578I06 gene
|
|
chr6_-_87699767
Show fit
|
20.29 |
ENSMUST00000238521.2
|
1810020O05Rik
|
Riken cDNA 1810020O05 gene
|
|
chr5_+_73808718
Show fit
|
20.20 |
ENSMUST00000071077.14
|
Spata18
|
spermatogenesis associated 18
|
|
chr5_+_98477157
Show fit
|
20.18 |
ENSMUST00000080333.8
|
Cfap299
|
cilia and flagella associated protein 299
|
|
chr3_-_37366567
Show fit
|
20.00 |
ENSMUST00000075537.7
ENSMUST00000071400.13
ENSMUST00000102955.11
ENSMUST00000140956.8
|
Cetn4
|
centrin 4
|
|
chr8_+_41128099
Show fit
|
19.94 |
ENSMUST00000051614.5
|
Adam24
|
a disintegrin and metallopeptidase domain 24 (testase 1)
|
|
chr4_+_100336003
Show fit
|
19.66 |
ENSMUST00000133493.9
ENSMUST00000092730.5
|
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative)
|
|
chr5_-_76478935
Show fit
|
19.64 |
ENSMUST00000122213.8
ENSMUST00000031145.7
|
Pdcl2
|
phosducin-like 2
|
|
chr7_+_90075762
Show fit
|
19.54 |
ENSMUST00000061391.9
|
Ccdc89
|
coiled-coil domain containing 89
|
|
chr19_-_23630143
Show fit
|
19.49 |
ENSMUST00000035849.5
|
1700028P14Rik
|
RIKEN cDNA 1700028P14 gene
|
|
chr1_-_53745920
Show fit
|
19.37 |
ENSMUST00000094964.7
|
Dnah7a
|
dynein, axonemal, heavy chain 7A
|
|
chr2_-_69619864
Show fit
|
18.90 |
ENSMUST00000094942.4
|
Ccdc173
|
coiled-coil domain containing 173
|
|
chr8_+_41246310
Show fit
|
18.65 |
ENSMUST00000056331.8
|
Adam20
|
a disintegrin and metallopeptidase domain 20
|
|
chrX_+_149981074
Show fit
|
18.61 |
ENSMUST00000184730.8
ENSMUST00000184392.8
ENSMUST00000096285.5
|
Wnk3
|
WNK lysine deficient protein kinase 3
|
|
chr4_+_125952349
Show fit
|
18.45 |
ENSMUST00000035497.5
ENSMUST00000143712.3
|
Oscp1
|
organic solute carrier partner 1
|
|
chr13_-_99121070
Show fit
|
18.37 |
ENSMUST00000054425.7
|
H2bl1
|
H2B.L histone variant 1
|
|
chr12_+_98886826
Show fit
|
18.00 |
ENSMUST00000085109.10
ENSMUST00000079146.13
|
Ttc8
|
tetratricopeptide repeat domain 8
|
|
chr18_-_34712123
Show fit
|
17.98 |
ENSMUST00000079287.12
|
Nme5
|
NME/NM23 family member 5
|
|
chr4_+_137989783
Show fit
|
17.80 |
ENSMUST00000105821.3
|
Kif17
|
kinesin family member 17
|
|
chr12_+_98594388
Show fit
|
17.76 |
ENSMUST00000048402.12
ENSMUST00000101144.10
ENSMUST00000101146.4
|
Spata7
|
spermatogenesis associated 7
|
|
chr12_+_57610897
Show fit
|
17.50 |
ENSMUST00000101398.10
|
Ttc6
|
tetratricopeptide repeat domain 6
|
|
chr1_-_131065967
Show fit
|
17.40 |
ENSMUST00000189756.2
|
Dyrk3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3
|
|
chr19_+_58931847
Show fit
|
17.22 |
ENSMUST00000054280.10
ENSMUST00000200910.4
|
Eno4
|
enolase 4
|
|
chr11_+_103540391
Show fit
|
17.00 |
ENSMUST00000057870.4
|
Rprml
|
reprimo-like
|
|
chr7_-_139263713
Show fit
|
16.86 |
ENSMUST00000129990.9
ENSMUST00000130453.8
|
Cfap46
|
cilia and flagella associated protein 46
|
|
chr6_-_113577606
Show fit
|
16.80 |
ENSMUST00000035870.5
|
Fancd2os
|
Fancd2 opposite strand
|
|
chr4_-_12087911
Show fit
|
16.64 |
ENSMUST00000050686.10
|
Tmem67
|
transmembrane protein 67
|
|
chr10_+_57521930
Show fit
|
16.58 |
ENSMUST00000177325.8
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific
|
|
chr4_+_41569775
Show fit
|
16.53 |
ENSMUST00000102963.10
|
Dnai1
|
dynein axonemal intermediate chain 1
|
|
chr3_+_154369496
Show fit
|
16.39 |
ENSMUST00000098496.9
|
Erich3
|
glutamate rich 3
|
|
chrX_+_56454507
Show fit
|
16.35 |
ENSMUST00000114725.3
|
Tm9sf5
|
transmembrane 9 superfamily member 5
|
|
chr11_+_11414256
Show fit
|
16.33 |
ENSMUST00000020410.11
|
Spata48
|
spermatogenesis associated 48
|
|
chr5_+_124802149
Show fit
|
16.13 |
ENSMUST00000141137.7
ENSMUST00000058440.12
|
Dnah10
|
dynein, axonemal, heavy chain 10
|
|
chr1_-_74974707
Show fit
|
15.84 |
ENSMUST00000094844.4
|
Cfap65
|
cilia and flagella associated protein 65
|
|
chr7_+_16693604
Show fit
|
15.80 |
ENSMUST00000038163.8
|
Pnmal1
|
PNMA-like 1
|
|
chr8_+_123939596
Show fit
|
15.73 |
ENSMUST00000212892.2
ENSMUST00000212161.2
ENSMUST00000060133.8
ENSMUST00000212346.2
ENSMUST00000212637.2
|
Spata33
|
spermatogenesis associated 33
|
|
chr9_+_110306052
Show fit
|
15.65 |
ENSMUST00000197248.5
ENSMUST00000061155.12
ENSMUST00000198043.5
ENSMUST00000084952.8
|
Kif9
|
kinesin family member 9
|
|
chr14_-_32110312
Show fit
|
15.53 |
ENSMUST00000100723.4
|
1700024G13Rik
|
RIKEN cDNA 1700024G13 gene
|
|
chr5_+_66417233
Show fit
|
15.53 |
ENSMUST00000202994.4
ENSMUST00000201100.4
|
Nsun7
|
NOL1/NOP2/Sun domain family, member 7
|
|
chr8_+_41205245
Show fit
|
15.39 |
ENSMUST00000096663.5
|
Adam25
|
a disintegrin and metallopeptidase domain 25 (testase 2)
|
|
chr1_+_74640590
Show fit
|
15.38 |
ENSMUST00000087183.11
ENSMUST00000148456.8
ENSMUST00000113694.8
|
Stk36
|
serine/threonine kinase 36
|
|
chr7_-_109038282
Show fit
|
15.36 |
ENSMUST00000106745.9
ENSMUST00000090414.11
ENSMUST00000141210.2
|
Stk33
|
serine/threonine kinase 33
|
|
chr12_-_81615248
Show fit
|
15.34 |
ENSMUST00000008582.4
|
Adam21
|
a disintegrin and metallopeptidase domain 21
|
|
chr2_+_152804405
Show fit
|
15.33 |
ENSMUST00000099197.9
ENSMUST00000103155.10
|
Ttll9
|
tubulin tyrosine ligase-like family, member 9
|
|
chr10_-_92917458
Show fit
|
15.26 |
ENSMUST00000212902.2
ENSMUST00000168617.3
ENSMUST00000168110.8
ENSMUST00000020200.14
|
Cfap54
|
cilia and flagella associated protein 54
|
|
chr11_-_105347500
Show fit
|
15.07 |
ENSMUST00000049995.10
ENSMUST00000100332.4
|
Marchf10
|
membrane associated ring-CH-type finger 10
|
|
chr17_-_5468938
Show fit
|
14.97 |
ENSMUST00000189788.2
|
Ldhal6b
|
lactate dehydrogenase A-like 6B
|
|
chr8_-_55177510
Show fit
|
14.87 |
ENSMUST00000175915.8
|
Wdr17
|
WD repeat domain 17
|
|
chr18_+_52748978
Show fit
|
14.79 |
ENSMUST00000072666.4
ENSMUST00000209270.2
|
Zfp474
|
zinc finger protein 474
|
|
chr4_+_155493668
Show fit
|
14.70 |
ENSMUST00000123952.9
ENSMUST00000238423.2
ENSMUST00000238620.2
ENSMUST00000151083.8
|
Cfap74
|
cilia and flagella associated protein 74
|
|
chr12_+_87247297
Show fit
|
14.58 |
ENSMUST00000182869.2
|
Samd15
|
sterile alpha motif domain containing 15
|
|
chr15_+_85016261
Show fit
|
14.42 |
ENSMUST00000023067.4
|
Ribc2
|
RIB43A domain with coiled-coils 2
|
|
chr11_+_54205722
Show fit
|
14.36 |
ENSMUST00000072178.11
ENSMUST00000101211.9
ENSMUST00000101213.9
ENSMUST00000064690.10
ENSMUST00000108899.8
|
Acsl6
|
acyl-CoA synthetase long-chain family member 6
|
|
chr17_-_65847731
Show fit
|
14.30 |
ENSMUST00000233117.2
ENSMUST00000062161.7
|
Tmem232
|
transmembrane protein 232
|
|
chr14_+_40794817
Show fit
|
14.24 |
ENSMUST00000189865.7
|
Dydc1
|
DPY30 domain containing 1
|
|
chr5_+_123390149
Show fit
|
14.06 |
ENSMUST00000121964.8
|
Wdr66
|
WD repeat domain 66
|
|
chr8_+_27937691
Show fit
|
13.96 |
ENSMUST00000081321.5
|
Poteg
|
POTE ankyrin domain family, member G
|
|
chr8_+_41276027
Show fit
|
13.94 |
ENSMUST00000066814.7
|
Adam39
|
a disintegrin and metallopeptidase domain 39
|
|
chr15_-_85016150
Show fit
|
13.79 |
ENSMUST00000023068.8
|
Smc1b
|
structural maintenance of chromosomes 1B
|
|
chr2_+_173364379
Show fit
|
13.61 |
ENSMUST00000029023.4
|
1700021F07Rik
|
RIKEN cDNA 1700021F07 gene
|
|
chr7_+_4925781
Show fit
|
13.60 |
ENSMUST00000207527.2
ENSMUST00000207687.2
ENSMUST00000208754.2
|
Nat14
|
N-acetyltransferase 14
|
|
chr9_+_22322875
Show fit
|
13.52 |
ENSMUST00000214436.2
|
9530077C05Rik
|
RIKEN cDNA 9530077C05 gene
|
|
chr7_-_83444050
Show fit
|
13.45 |
ENSMUST00000011298.14
|
Cfap161
|
cilia and flagella associated protein 161
|
|
chr7_-_101582987
Show fit
|
13.44 |
ENSMUST00000106964.8
ENSMUST00000106963.2
ENSMUST00000078448.11
ENSMUST00000106966.8
|
Lrrc51
|
leucine rich repeat containing 51
|
|
chr2_-_140231618
Show fit
|
13.37 |
ENSMUST00000122367.8
ENSMUST00000120133.2
|
Sel1l2
|
sel-1 suppressor of lin-12-like 2 (C. elegans)
|
|
chr11_+_62952431
Show fit
|
13.37 |
ENSMUST00000035732.5
|
Tekt3
|
tektin 3
|
|
chr12_+_84161095
Show fit
|
13.28 |
ENSMUST00000123491.8
ENSMUST00000046340.9
ENSMUST00000136159.2
|
Dnal1
|
dynein, axonemal, light chain 1
|
|
chr1_-_75187417
Show fit
|
13.27 |
ENSMUST00000113623.8
|
Glb1l
|
galactosidase, beta 1-like
|
|
chr2_-_32665596
Show fit
|
13.14 |
ENSMUST00000161430.8
|
Ttc16
|
tetratricopeptide repeat domain 16
|
|
chr17_+_71980249
Show fit
|
13.05 |
ENSMUST00000097284.10
|
Togaram2
|
TOG array regulator of axonemal microtubules 2
|
|
chr8_-_13612397
Show fit
|
13.01 |
ENSMUST00000187391.7
ENSMUST00000134023.9
ENSMUST00000151400.10
|
1700029H14Rik
|
RIKEN cDNA 1700029H14 gene
|
|
chr6_+_30509826
Show fit
|
12.72 |
ENSMUST00000031797.11
|
Ssmem1
|
serine-rich single-pass membrane protein 1
|
|
chr7_+_99508318
Show fit
|
12.57 |
ENSMUST00000036155.10
|
Xrra1
|
X-ray radiation resistance associated 1
|
|
chr15_+_51903503
Show fit
|
12.49 |
ENSMUST00000090025.5
|
Aard
|
alanine and arginine rich domain containing protein
|
|
chr10_+_61007733
Show fit
|
12.48 |
ENSMUST00000122261.8
ENSMUST00000121297.8
ENSMUST00000035894.12
|
Tbata
|
thymus, brain and testes associated
|
|
chr6_+_29471436
Show fit
|
12.46 |
ENSMUST00000171317.2
|
Atp6v1fnb
|
Atp6v1f neighbor
|
|
chr18_+_74416161
Show fit
|
12.41 |
ENSMUST00000114895.4
|
Cfap53
|
cilia and flagella associated protein 53
|
|
chr12_-_76293459
Show fit
|
12.31 |
ENSMUST00000219327.2
ENSMUST00000021453.6
ENSMUST00000218426.2
|
Tex21
|
testis expressed gene 21
|
|
chr10_-_22607136
Show fit
|
12.24 |
ENSMUST00000238910.2
ENSMUST00000127698.8
|
ENSMUSG00000118528.2
Tbpl1
|
novel protein
TATA box binding protein-like 1
|
|
chr4_-_129508047
Show fit
|
12.02 |
ENSMUST00000106037.3
ENSMUST00000179209.8
|
Dcdc2b
|
doublecortin domain containing 2b
|
|
chr11_-_94412297
Show fit
|
11.93 |
ENSMUST00000239238.2
|
Mycbpap
|
MYCBP associated protein
|
|
chr10_+_57521958
Show fit
|
11.81 |
ENSMUST00000177473.8
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific
|
|
chr1_-_75187441
Show fit
|
11.71 |
ENSMUST00000185448.2
|
Glb1l
|
galactosidase, beta 1-like
|
|
chr11_+_51895018
Show fit
|
11.70 |
ENSMUST00000109079.9
ENSMUST00000063303.11
ENSMUST00000109077.9
ENSMUST00000109080.9
ENSMUST00000109081.9
ENSMUST00000121591.8
ENSMUST00000109078.8
ENSMUST00000063321.13
ENSMUST00000135076.8
ENSMUST00000120374.8
|
Cdkl3
|
cyclin-dependent kinase-like 3
|
|
chr3_-_105921946
Show fit
|
11.68 |
ENSMUST00000010280.11
|
Pifo
|
primary cilia formation
|
|
chr3_-_75389047
Show fit
|
11.68 |
ENSMUST00000193989.4
ENSMUST00000203169.3
|
Wdr49
|
WD repeat domain 49
|
|
chr19_-_28945194
Show fit
|
11.66 |
ENSMUST00000162110.8
|
Spata6l
|
spermatogenesis associated 6 like
|
|
chr14_+_40794893
Show fit
|
11.62 |
ENSMUST00000161837.2
|
Dydc1
|
DPY30 domain containing 1
|
|
chr1_-_63769465
Show fit
|
11.51 |
ENSMUST00000114094.3
|
Mdh1b
|
malate dehydrogenase 1B, NAD (soluble)
|
|
chr17_-_31496301
Show fit
|
11.49 |
ENSMUST00000235144.2
|
Rsph1
|
radial spoke head 1 homolog (Chlamydomonas)
|
|
chr7_+_119495058
Show fit
|
11.39 |
ENSMUST00000106518.9
ENSMUST00000207270.2
ENSMUST00000208424.2
ENSMUST00000208202.2
ENSMUST00000054440.11
|
Lyrm1
|
LYR motif containing 1
|
|
chr8_-_106434565
Show fit
|
11.39 |
ENSMUST00000013299.11
|
Enkd1
|
enkurin domain containing 1
|
|
chr19_-_40600619
Show fit
|
11.38 |
ENSMUST00000132452.2
ENSMUST00000135795.8
ENSMUST00000025981.15
|
Tctn3
|
tectonic family member 3
|
|
chr7_+_16678568
Show fit
|
11.36 |
ENSMUST00000094807.6
|
Pnmal2
|
PNMA-like 2
|
|
chr8_+_55053809
Show fit
|
11.23 |
ENSMUST00000033917.7
|
Spata4
|
spermatogenesis associated 4
|
|
chr11_+_102556397
Show fit
|
11.08 |
ENSMUST00000100378.4
|
Meioc
|
meiosis specific with coiled-coil domain
|
|
chr12_-_98225676
Show fit
|
10.99 |
ENSMUST00000021390.9
|
Galc
|
galactosylceramidase
|
|
chrX_-_17185555
Show fit
|
10.91 |
ENSMUST00000026014.8
|
Efhc2
|
EF-hand domain (C-terminal) containing 2
|
|
chr4_+_117109204
Show fit
|
10.87 |
ENSMUST00000125943.8
ENSMUST00000106434.8
|
Tmem53
|
transmembrane protein 53
|
|
chr17_+_11059248
Show fit
|
10.84 |
ENSMUST00000191124.7
|
Prkn
|
parkin RBR E3 ubiquitin protein ligase
|
|
chr10_+_74872898
Show fit
|
10.82 |
ENSMUST00000147802.9
ENSMUST00000020391.13
ENSMUST00000234625.2
|
Rab36
|
RAB36, member RAS oncogene family
|
|
chr5_+_120727068
Show fit
|
10.81 |
ENSMUST00000069259.9
ENSMUST00000094391.6
|
Iqcd
|
IQ motif containing D
|
|
chr17_+_80597632
Show fit
|
10.74 |
ENSMUST00000227729.2
ENSMUST00000061703.10
|
Morn2
|
MORN repeat containing 2
|
|
chr5_+_143869875
Show fit
|
10.70 |
ENSMUST00000166847.8
|
Rsph10b
|
radial spoke head 10 homolog B (Chlamydomonas)
|
|
chr17_-_31496352
Show fit
|
10.68 |
ENSMUST00000024832.9
|
Rsph1
|
radial spoke head 1 homolog (Chlamydomonas)
|
|
chr15_+_75881712
Show fit
|
10.59 |
ENSMUST00000187868.3
|
Iqank1
|
IQ motif and ankyrin repeat containing 1
|
|
chr1_-_4479445
Show fit
|
10.56 |
ENSMUST00000208660.2
|
Rp1
|
retinitis pigmentosa 1 (human)
|
|
chr17_-_65847777
Show fit
|
10.52 |
ENSMUST00000086722.10
|
Tmem232
|
transmembrane protein 232
|
|
chr1_+_60448703
Show fit
|
10.37 |
ENSMUST00000052332.15
|
Abi2
|
abl interactor 2
|
|
chr3_-_75072319
Show fit
|
10.29 |
ENSMUST00000124618.2
|
Zbbx
|
zinc finger, B-box domain containing
|
|
chr12_-_11485639
Show fit
|
10.28 |
ENSMUST00000220506.2
|
Vsnl1
|
visinin-like 1
|
|
chr9_-_63509699
Show fit
|
10.28 |
ENSMUST00000171243.2
ENSMUST00000163982.8
ENSMUST00000163624.8
|
Iqch
|
IQ motif containing H
|
|
chr7_-_109037992
Show fit
|
10.26 |
ENSMUST00000121748.8
|
Stk33
|
serine/threonine kinase 33
|
|
chr3_-_50398027
Show fit
|
10.25 |
ENSMUST00000029297.6
ENSMUST00000194462.6
|
Slc7a11
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11
|
|
chr2_-_25104628
Show fit
|
10.24 |
ENSMUST00000043774.11
ENSMUST00000114363.2
|
Stpg3
|
sperm tail PG rich repeat containing 3
|
|
chr15_-_89476057
Show fit
|
10.20 |
ENSMUST00000094056.12
ENSMUST00000023294.15
|
Rabl2
|
RAB, member RAS oncogene family-like 2
|
|
chr11_+_119833589
Show fit
|
10.13 |
ENSMUST00000106231.8
ENSMUST00000075180.12
ENSMUST00000103021.10
ENSMUST00000026436.10
ENSMUST00000106233.2
|
Baiap2
|
brain-specific angiogenesis inhibitor 1-associated protein 2
|
|
chr4_-_41569500
Show fit
|
10.11 |
ENSMUST00000108049.9
ENSMUST00000108052.10
ENSMUST00000108050.2
|
Fam219a
|
family with sequence similarity 219, member A
|
|
chr4_+_117109148
Show fit
|
9.99 |
ENSMUST00000062824.12
|
Tmem53
|
transmembrane protein 53
|
|
chr2_+_29780073
Show fit
|
9.98 |
ENSMUST00000028128.13
|
Odf2
|
outer dense fiber of sperm tails 2
|
|
chr1_+_60448813
Show fit
|
9.97 |
ENSMUST00000188594.7
ENSMUST00000188618.7
ENSMUST00000189980.7
|
Abi2
|
abl interactor 2
|
|
chr4_+_123077286
Show fit
|
9.89 |
ENSMUST00000126995.2
|
Hpcal4
|
hippocalcin-like 4
|
|
chr7_-_109038202
Show fit
|
9.89 |
ENSMUST00000121378.2
|
Stk33
|
serine/threonine kinase 33
|
|
chr8_-_44029744
Show fit
|
9.84 |
ENSMUST00000049577.3
|
Adam26a
|
a disintegrin and metallopeptidase domain 26A (testase 3)
|
|
chr1_-_90081118
Show fit
|
9.84 |
ENSMUST00000113094.9
|
Iqca
|
IQ motif containing with AAA domain
|
|
chr4_-_128856213
Show fit
|
9.82 |
ENSMUST00000119354.8
ENSMUST00000106068.8
ENSMUST00000030581.10
|
Azin2
|
antizyme inhibitor 2
|
|
chr14_+_57661514
Show fit
|
9.80 |
ENSMUST00000122063.8
|
Ift88
|
intraflagellar transport 88
|
|
chr19_-_58932026
Show fit
|
9.79 |
ENSMUST00000237297.2
|
Hspa12a
|
heat shock protein 12A
|
|
chr7_+_119495515
Show fit
|
9.76 |
ENSMUST00000106517.9
|
Lyrm1
|
LYR motif containing 1
|
|
chr2_-_111059901
Show fit
|
9.75 |
ENSMUST00000028577.3
|
A26c3
|
ANKRD26-like family C, member 3
|