Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Rhox11
Z-value: 0.54
Transcription factors associated with Rhox11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rhox11
|
ENSMUSG00000051038.11 | Rhox11 |
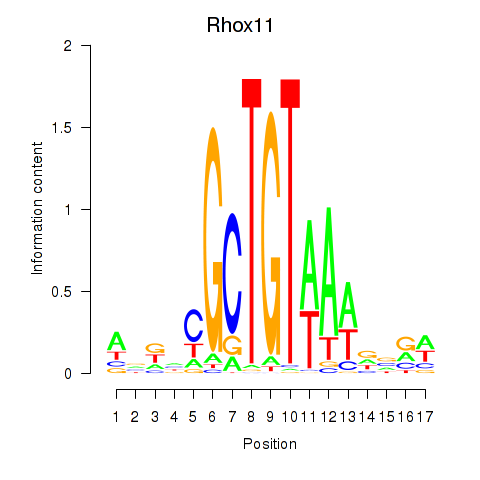
Activity profile of Rhox11 motif
Sorted Z-values of Rhox11 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Rhox11
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_110867807 | 4.41 |
ENSMUST00000197575.2
|
Ltf
|
lactotransferrin |
| chr19_+_11724913 | 3.88 |
ENSMUST00000025585.4
|
Cblif
|
cobalamin binding intrinsic factor |
| chr8_+_106554353 | 2.42 |
ENSMUST00000212566.2
ENSMUST00000034365.5 |
Tsnaxip1
|
translin-associated factor X (Tsnax) interacting protein 1 |
| chr3_-_92393193 | 2.31 |
ENSMUST00000054599.8
|
Sprr1a
|
small proline-rich protein 1A |
| chr3_+_90511068 | 2.11 |
ENSMUST00000001046.7
|
S100a4
|
S100 calcium binding protein A4 |
| chr5_+_98477157 | 1.60 |
ENSMUST00000080333.8
|
Cfap299
|
cilia and flagella associated protein 299 |
| chr18_-_42084249 | 1.48 |
ENSMUST00000070949.6
ENSMUST00000235606.2 |
Prelid2
|
PRELI domain containing 2 |
| chr3_-_105908789 | 1.42 |
ENSMUST00000066319.8
|
Pifo
|
primary cilia formation |
| chr8_+_105558204 | 1.37 |
ENSMUST00000059449.7
|
Ces2b
|
carboxyesterase 2B |
| chr5_+_122239030 | 1.37 |
ENSMUST00000139213.8
ENSMUST00000111751.8 ENSMUST00000155612.8 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr10_+_102376109 | 1.36 |
ENSMUST00000055355.6
|
Rassf9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr4_-_119241024 | 1.34 |
ENSMUST00000127149.8
ENSMUST00000152879.9 ENSMUST00000238673.2 ENSMUST00000238485.2 |
Ccdc30
|
coiled-coil domain containing 30 |
| chr9_-_14651985 | 1.31 |
ENSMUST00000076946.4
ENSMUST00000115644.10 |
Piwil4
|
piwi-like RNA-mediated gene silencing 4 |
| chr12_-_30423356 | 1.29 |
ENSMUST00000021004.14
|
Sntg2
|
syntrophin, gamma 2 |
| chr17_+_41121979 | 1.28 |
ENSMUST00000024721.8
ENSMUST00000233740.2 |
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr10_+_58230203 | 1.26 |
ENSMUST00000105468.2
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr12_+_102521225 | 1.18 |
ENSMUST00000021610.7
|
Chga
|
chromogranin A |
| chr12_+_104067346 | 1.15 |
ENSMUST00000021495.4
|
Serpina5
|
serine (or cysteine) peptidase inhibitor, clade A, member 5 |
| chr7_+_43361930 | 1.14 |
ENSMUST00000066834.8
|
Klk13
|
kallikrein related-peptidase 13 |
| chr9_-_107167046 | 1.13 |
ENSMUST00000035194.8
|
Mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr5_+_37892863 | 1.12 |
ENSMUST00000073554.4
|
Cytl1
|
cytokine-like 1 |
| chr6_-_129428869 | 1.06 |
ENSMUST00000203162.3
|
Clec1a
|
C-type lectin domain family 1, member a |
| chr4_-_119240885 | 1.05 |
ENSMUST00000238422.2
ENSMUST00000238719.2 ENSMUST00000238723.2 ENSMUST00000044781.9 ENSMUST00000084307.5 ENSMUST00000148236.9 ENSMUST00000127474.3 ENSMUST00000238704.2 |
Ccdc30
|
coiled-coil domain containing 30 |
| chr7_+_46633328 | 1.00 |
ENSMUST00000070660.11
ENSMUST00000188312.2 |
Gm9999
|
predicted gene 9999 |
| chr8_+_73072877 | 1.00 |
ENSMUST00000067912.8
|
Klf2
|
Kruppel-like factor 2 (lung) |
| chr1_+_58152295 | 0.99 |
ENSMUST00000040999.14
ENSMUST00000162011.3 |
Aox3
|
aldehyde oxidase 3 |
| chr6_+_78382131 | 0.97 |
ENSMUST00000023906.4
|
Reg2
|
regenerating islet-derived 2 |
| chr18_-_60635059 | 0.95 |
ENSMUST00000042710.8
|
Smim3
|
small integral membrane protein 3 |
| chr10_+_58230183 | 0.92 |
ENSMUST00000020077.11
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr14_-_75829389 | 0.91 |
ENSMUST00000165569.3
ENSMUST00000035243.5 |
Cby2
|
chibby family member 2 |
| chr4_+_62398262 | 0.91 |
ENSMUST00000030088.12
ENSMUST00000107449.4 |
Bspry
|
B-box and SPRY domain containing |
| chr9_-_107166543 | 0.90 |
ENSMUST00000192054.2
|
Mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr12_-_56583582 | 0.89 |
ENSMUST00000001536.9
|
Nkx2-1
|
NK2 homeobox 1 |
| chr16_-_57575070 | 0.88 |
ENSMUST00000089332.5
|
Col8a1
|
collagen, type VIII, alpha 1 |
| chr11_+_23234644 | 0.87 |
ENSMUST00000150750.3
|
Xpo1
|
exportin 1 |
| chr17_+_79922329 | 0.86 |
ENSMUST00000040368.3
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr12_-_99849660 | 0.85 |
ENSMUST00000221929.2
ENSMUST00000046485.5 |
Efcab11
|
EF-hand calcium binding domain 11 |
| chr6_+_139564196 | 0.83 |
ENSMUST00000188066.2
ENSMUST00000190962.7 |
Pik3c2g
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
| chr10_+_41179966 | 0.80 |
ENSMUST00000173494.4
|
Ak9
|
adenylate kinase 9 |
| chr8_-_106553822 | 0.80 |
ENSMUST00000239468.2
ENSMUST00000041400.6 |
Ranbp10
|
RAN binding protein 10 |
| chr5_-_82271183 | 0.79 |
ENSMUST00000186079.2
ENSMUST00000185607.2 |
1700031L13Rik
|
RIKEN cDNA 1700031L13 gene |
| chr4_-_119241002 | 0.78 |
ENSMUST00000238721.2
|
Ccdc30
|
coiled-coil domain containing 30 |
| chr3_-_122828592 | 0.76 |
ENSMUST00000029761.14
|
Myoz2
|
myozenin 2 |
| chr1_-_182576739 | 0.75 |
ENSMUST00000060041.7
|
Ccdc185
|
coiled-coil domain containing 185 |
| chrX_-_78560891 | 0.74 |
ENSMUST00000197180.6
ENSMUST00000101410.3 |
Cfap47
|
cilia and flagella associated protein 47 |
| chr1_+_153628598 | 0.67 |
ENSMUST00000182538.3
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent) |
| chr6_-_129600798 | 0.67 |
ENSMUST00000095412.10
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr6_-_129600812 | 0.66 |
ENSMUST00000168919.8
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr14_-_21898992 | 0.65 |
ENSMUST00000124549.9
|
Comtd1
|
catechol-O-methyltransferase domain containing 1 |
| chr9_+_106377181 | 0.63 |
ENSMUST00000085114.8
|
Iqcf1
|
IQ motif containing F1 |
| chr13_-_95661726 | 0.60 |
ENSMUST00000022185.10
|
F2rl1
|
coagulation factor II (thrombin) receptor-like 1 |
| chr8_+_126456710 | 0.60 |
ENSMUST00000143504.8
|
Ntpcr
|
nucleoside-triphosphatase, cancer-related |
| chr7_+_126184108 | 0.60 |
ENSMUST00000039522.8
|
Apobr
|
apolipoprotein B receptor |
| chr6_-_129637519 | 0.58 |
ENSMUST00000119533.2
ENSMUST00000145984.8 ENSMUST00000118401.8 ENSMUST00000112057.9 ENSMUST00000071920.11 |
Klrc2
|
killer cell lectin-like receptor subfamily C, member 2 |
| chr11_+_29323618 | 0.57 |
ENSMUST00000040182.13
ENSMUST00000109477.2 |
Ccdc88a
|
coiled coil domain containing 88A |
| chr5_+_7354113 | 0.56 |
ENSMUST00000088796.3
|
Tex47
|
testis expressed 47 |
| chr3_+_127584449 | 0.55 |
ENSMUST00000171621.3
|
Tifa
|
TRAF-interacting protein with forkhead-associated domain |
| chr5_+_143869875 | 0.54 |
ENSMUST00000166847.8
|
Rsph10b
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr5_+_21391282 | 0.53 |
ENSMUST00000036031.13
ENSMUST00000198937.2 |
Gsap
|
gamma-secretase activating protein |
| chr11_-_103505565 | 0.52 |
ENSMUST00000167262.2
|
Gm884
|
predicted gene 884 |
| chr5_+_7354130 | 0.51 |
ENSMUST00000160634.2
ENSMUST00000159546.2 |
Tex47
|
testis expressed 47 |
| chr11_-_69786324 | 0.51 |
ENSMUST00000001631.7
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr4_+_126450728 | 0.51 |
ENSMUST00000048391.15
|
Clspn
|
claspin |
| chr4_+_138926577 | 0.50 |
ENSMUST00000145368.8
|
Capzb
|
capping protein (actin filament) muscle Z-line, beta |
| chr5_+_43976218 | 0.50 |
ENSMUST00000101237.8
|
Bst1
|
bone marrow stromal cell antigen 1 |
| chr11_-_97673203 | 0.50 |
ENSMUST00000128801.2
ENSMUST00000103146.5 |
Rpl23
|
ribosomal protein L23 |
| chr1_+_180769890 | 0.47 |
ENSMUST00000161847.8
|
Tmem63a
|
transmembrane protein 63a |
| chr15_+_31225302 | 0.45 |
ENSMUST00000186425.7
|
Dap
|
death-associated protein |
| chr3_-_30654012 | 0.45 |
ENSMUST00000047630.7
|
Actrt3
|
actin related protein T3 |
| chr7_-_30364394 | 0.43 |
ENSMUST00000019697.9
|
Haus5
|
HAUS augmin-like complex, subunit 5 |
| chrX_-_101200670 | 0.39 |
ENSMUST00000056904.3
|
Ercc6l
|
excision repair cross-complementing rodent repair deficiency complementation group 6 like |
| chr10_-_29411857 | 0.39 |
ENSMUST00000092623.5
|
Rspo3
|
R-spondin 3 |
| chr5_-_87682972 | 0.38 |
ENSMUST00000120150.2
|
Sult1b1
|
sulfotransferase family 1B, member 1 |
| chr3_-_49711706 | 0.35 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chr7_+_126184141 | 0.32 |
ENSMUST00000137646.8
|
Apobr
|
apolipoprotein B receptor |
| chr3_-_49711765 | 0.31 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr15_-_38079089 | 0.30 |
ENSMUST00000110336.4
|
Ubr5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr6_+_54241830 | 0.29 |
ENSMUST00000146114.8
|
Chn2
|
chimerin 2 |
| chr13_-_3661064 | 0.25 |
ENSMUST00000096069.5
|
Tasor2
|
transcription activation suppressor family member 2 |
| chr9_-_20432562 | 0.24 |
ENSMUST00000215908.2
ENSMUST00000068296.8 ENSMUST00000174462.8 ENSMUST00000213418.2 |
Zfp266
|
zinc finger protein 266 |
| chr14_-_30890544 | 0.22 |
ENSMUST00000036618.14
|
Stab1
|
stabilin 1 |
| chr11_+_70529944 | 0.21 |
ENSMUST00000055184.7
ENSMUST00000108551.3 |
Gp1ba
|
glycoprotein 1b, alpha polypeptide |
| chr11_-_97591150 | 0.21 |
ENSMUST00000018681.14
|
Pcgf2
|
polycomb group ring finger 2 |
| chr1_+_40844739 | 0.18 |
ENSMUST00000114765.4
|
Tmem182
|
transmembrane protein 182 |
| chr3_-_51184730 | 0.17 |
ENSMUST00000195432.2
ENSMUST00000091144.11 ENSMUST00000156983.3 |
Elf2
|
E74-like factor 2 |
| chr3_-_51184895 | 0.16 |
ENSMUST00000108051.8
ENSMUST00000108053.9 |
Elf2
|
E74-like factor 2 |
| chr14_-_20546848 | 0.16 |
ENSMUST00000022353.5
|
Mss51
|
MSS51 mitochondrial translational activator |
| chr7_+_19927635 | 0.16 |
ENSMUST00000168984.2
|
Vmn1r95
|
vomeronasal 1 receptor, 95 |
| chr10_-_61109188 | 0.16 |
ENSMUST00000092486.11
|
Adamts14
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 14 |
| chr11_-_5331734 | 0.15 |
ENSMUST00000172492.8
|
Znrf3
|
zinc and ring finger 3 |
| chrX_+_104123341 | 0.15 |
ENSMUST00000033577.11
|
Pbdc1
|
polysaccharide biosynthesis domain containing 1 |
| chr2_+_35512172 | 0.15 |
ENSMUST00000112992.9
|
Dab2ip
|
disabled 2 interacting protein |
| chr17_-_46558894 | 0.14 |
ENSMUST00000142706.9
ENSMUST00000173349.8 ENSMUST00000087026.13 |
Polr1c
|
polymerase (RNA) I polypeptide C |
| chr12_+_76884182 | 0.14 |
ENSMUST00000041008.10
|
Fntb
|
farnesyltransferase, CAAX box, beta |
| chr15_+_25742397 | 0.13 |
ENSMUST00000135981.8
|
Myo10
|
myosin X |
| chr15_+_91722458 | 0.12 |
ENSMUST00000109277.8
|
Smgc
|
submandibular gland protein C |
| chr10_-_77845571 | 0.12 |
ENSMUST00000020522.9
|
Pfkl
|
phosphofructokinase, liver, B-type |
| chr6_+_121815473 | 0.12 |
ENSMUST00000032228.9
|
Mug1
|
murinoglobulin 1 |
| chr13_+_49806772 | 0.11 |
ENSMUST00000223264.2
ENSMUST00000221142.2 ENSMUST00000222333.2 ENSMUST00000021824.8 |
Nol8
|
nucleolar protein 8 |
| chr2_-_57003064 | 0.11 |
ENSMUST00000112627.2
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr15_+_91722524 | 0.11 |
ENSMUST00000109276.8
ENSMUST00000088555.10 ENSMUST00000100293.9 ENSMUST00000126508.8 ENSMUST00000239545.1 |
Smgc
ENSMUSG00000118670.1
|
submandibular gland protein C mucin 19 |
| chr1_+_132433968 | 0.11 |
ENSMUST00000058167.3
|
Tmem81
|
transmembrane protein 81 |
| chr8_+_89373943 | 0.11 |
ENSMUST00000118370.8
ENSMUST00000054324.15 |
Nod2
|
nucleotide-binding oligomerization domain containing 2 |
| chr13_+_95461703 | 0.10 |
ENSMUST00000045909.8
|
Zbed3
|
zinc finger, BED type containing 3 |
| chr9_-_20556031 | 0.10 |
ENSMUST00000148631.8
ENSMUST00000131128.2 ENSMUST00000151861.9 ENSMUST00000131343.8 ENSMUST00000086458.10 |
Fbxl12
|
F-box and leucine-rich repeat protein 12 |
| chr16_+_20470402 | 0.10 |
ENSMUST00000007212.9
ENSMUST00000232629.2 |
Psmd2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chr12_+_105113292 | 0.09 |
ENSMUST00000081379.7
|
Tcl1b2
|
T cell leukemia/lymphoma 1B, 2 |
| chr8_+_105673981 | 0.09 |
ENSMUST00000076384.6
ENSMUST00000212926.2 |
Ces2f
|
carboxylesterase 2F |
| chr7_-_103214484 | 0.07 |
ENSMUST00000106886.2
|
Olfr616
|
olfactory receptor 616 |
| chr7_+_102549549 | 0.07 |
ENSMUST00000098215.3
|
Olfr570
|
olfactory receptor 570 |
| chr13_+_41040657 | 0.06 |
ENSMUST00000069958.15
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr13_+_22477363 | 0.05 |
ENSMUST00000091735.2
|
Vmn1r196
|
vomeronasal 1 receptor 196 |
| chr16_-_88609108 | 0.04 |
ENSMUST00000232664.2
|
Gm20741
|
predicted gene, 20741 |
| chr15_-_79658608 | 0.04 |
ENSMUST00000229644.2
ENSMUST00000023055.8 |
Dnal4
|
dynein, axonemal, light chain 4 |
| chr9_-_50679408 | 0.04 |
ENSMUST00000177546.8
ENSMUST00000176238.2 |
1110032A03Rik
|
RIKEN cDNA 1110032A03 gene |
| chr15_-_79658584 | 0.03 |
ENSMUST00000069877.12
|
Dnal4
|
dynein, axonemal, light chain 4 |
| chr7_-_44773750 | 0.02 |
ENSMUST00000211725.2
ENSMUST00000003521.10 |
Rps11
|
ribosomal protein S11 |
| chr18_-_70377653 | 0.02 |
ENSMUST00000025390.4
|
Dynap
|
dynactin associated protein |
| chr4_-_140867038 | 0.02 |
ENSMUST00000148204.8
ENSMUST00000102487.4 |
Szrd1
|
SUZ RNA binding domain containing 1 |
| chr14_-_51311892 | 0.01 |
ENSMUST00000216202.2
|
Olfr750
|
olfactory receptor 750 |
| chr7_-_141794815 | 0.01 |
ENSMUST00000211591.2
|
Gm40460
|
predicted gene, 40460 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.4 | GO:1900191 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.5 | 1.4 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.4 | 1.3 | GO:0030887 | positive regulation of myeloid dendritic cell activation(GO:0030887) |
| 0.3 | 1.3 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.3 | 0.9 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.3 | 1.2 | GO:1901079 | positive regulation of relaxation of muscle(GO:1901079) regulation of dense core granule biogenesis(GO:2000705) |
| 0.2 | 0.6 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) positive regulation of renin secretion into blood stream(GO:1900135) positive regulation of eosinophil activation(GO:1902568) |
| 0.2 | 0.8 | GO:0046066 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) purine deoxyribonucleoside triphosphate biosynthetic process(GO:0009216) dGDP metabolic process(GO:0046066) |
| 0.2 | 1.0 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.2 | 2.2 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.2 | 2.0 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 1.0 | GO:0071499 | response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.1 | 0.5 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.1 | 0.6 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.1 | 0.6 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.9 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.3 | GO:1905077 | negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.1 | 0.4 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 1.3 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.1 | 2.3 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 1.1 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.5 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.1 | GO:0032499 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) detection of peptidoglycan(GO:0032499) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 1.0 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.1 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.7 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.5 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.0 | 0.8 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 1.5 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.4 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.5 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 0.1 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.1 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.4 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.2 | 1.3 | GO:0071547 | piP-body(GO:0071547) |
| 0.2 | 1.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 1.1 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.1 | 1.4 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 1.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.9 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 2.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.9 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.5 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.5 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.4 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 2.0 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0032394 | MHC class Ib receptor activity(GO:0032394) |
| 0.4 | 1.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.4 | 2.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.3 | 1.3 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.2 | 1.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.2 | 2.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.2 | 0.8 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.2 | 1.0 | GO:0004031 | aldehyde oxidase activity(GO:0004031) xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) molybdenum ion binding(GO:0030151) |
| 0.2 | 0.8 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.2 | 0.6 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 4.4 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 2.0 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.5 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.1 | 0.9 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 0.8 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 0.9 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 1.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.6 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.1 | 0.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.5 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.9 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.9 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.1 | 1.3 | GO:0022840 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.8 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 1.9 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 1.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.7 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.4 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.6 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 2.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.9 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 2.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.6 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.8 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.0 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 2.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.9 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 1.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |