Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Rora
Z-value: 1.00
Transcription factors associated with Rora
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rora
|
ENSMUSG00000032238.18 | Rora |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rora | mm39_v1_chr9_+_68561042_68561068 | -0.14 | 2.4e-01 | Click! |
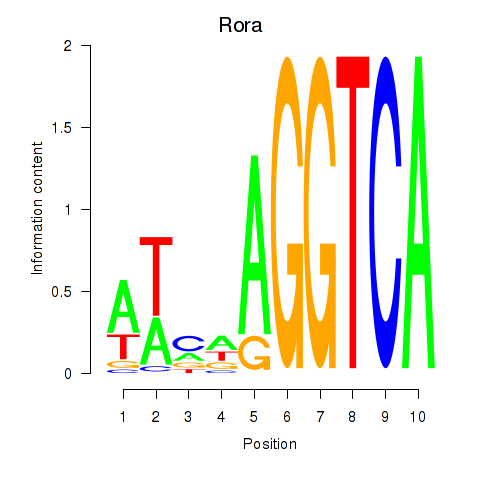
Activity profile of Rora motif
Sorted Z-values of Rora motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Rora
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_52165682 | 7.83 |
ENSMUST00000238914.2
|
Tcf7
|
transcription factor 7, T cell specific |
| chr15_-_42540363 | 7.81 |
ENSMUST00000022921.7
|
Angpt1
|
angiopoietin 1 |
| chr7_+_28140352 | 6.45 |
ENSMUST00000078845.13
|
Gmfg
|
glia maturation factor, gamma |
| chr15_-_66673425 | 6.24 |
ENSMUST00000168589.8
|
Sla
|
src-like adaptor |
| chr7_-_126014027 | 6.09 |
ENSMUST00000032968.7
ENSMUST00000206325.2 |
Cd19
|
CD19 antigen |
| chr2_+_131028861 | 5.82 |
ENSMUST00000028804.15
ENSMUST00000079857.9 |
Cdc25b
|
cell division cycle 25B |
| chr7_+_127728712 | 5.26 |
ENSMUST00000033053.8
ENSMUST00000205460.2 |
Itgax
|
integrin alpha X |
| chr3_-_83947416 | 5.03 |
ENSMUST00000192095.6
ENSMUST00000191758.6 ENSMUST00000052342.9 |
Tmem131l
|
transmembrane 131 like |
| chr10_+_84412490 | 4.60 |
ENSMUST00000020223.8
|
Tcp11l2
|
t-complex 11 (mouse) like 2 |
| chr7_+_28140450 | 4.41 |
ENSMUST00000135686.2
|
Gmfg
|
glia maturation factor, gamma |
| chr19_+_6450553 | 4.22 |
ENSMUST00000146831.8
ENSMUST00000035716.15 ENSMUST00000138555.8 ENSMUST00000167240.8 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr7_-_4525426 | 4.09 |
ENSMUST00000209148.2
ENSMUST00000098859.10 |
Tnni3
|
troponin I, cardiac 3 |
| chr7_-_100164007 | 4.04 |
ENSMUST00000207405.2
|
Dnajb13
|
DnaJ heat shock protein family (Hsp40) member B13 |
| chr17_+_8454939 | 3.98 |
ENSMUST00000164411.10
|
Ccr6
|
chemokine (C-C motif) receptor 6 |
| chr15_-_66432938 | 3.73 |
ENSMUST00000048372.7
|
Tmem71
|
transmembrane protein 71 |
| chr16_-_20245138 | 3.56 |
ENSMUST00000079158.13
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr10_-_99595498 | 3.50 |
ENSMUST00000056085.6
|
Csl
|
citrate synthase like |
| chr15_-_7427759 | 3.41 |
ENSMUST00000058593.10
|
Egflam
|
EGF-like, fibronectin type III and laminin G domains |
| chr15_-_7427815 | 3.40 |
ENSMUST00000096494.5
|
Egflam
|
EGF-like, fibronectin type III and laminin G domains |
| chr6_-_124698805 | 3.38 |
ENSMUST00000173315.8
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr11_+_4186391 | 3.34 |
ENSMUST00000075221.3
|
Osm
|
oncostatin M |
| chr7_-_4525793 | 3.28 |
ENSMUST00000140424.8
|
Tnni3
|
troponin I, cardiac 3 |
| chr11_+_116089678 | 3.27 |
ENSMUST00000021130.7
|
Ten1
|
TEN1 telomerase capping complex subunit |
| chr3_-_108797022 | 3.22 |
ENSMUST00000180063.8
ENSMUST00000053065.8 |
Fndc7
|
fibronectin type III domain containing 7 |
| chr4_+_131600918 | 3.17 |
ENSMUST00000053819.6
|
Srsf4
|
serine and arginine-rich splicing factor 4 |
| chr2_-_60711706 | 3.12 |
ENSMUST00000164147.8
ENSMUST00000112509.2 |
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr3_-_20329823 | 3.04 |
ENSMUST00000011607.6
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chrX_-_104919201 | 3.02 |
ENSMUST00000198209.2
|
Atrx
|
ATRX, chromatin remodeler |
| chr11_+_70548022 | 3.01 |
ENSMUST00000157027.8
ENSMUST00000072841.12 ENSMUST00000108548.8 ENSMUST00000126241.8 |
Eno3
|
enolase 3, beta muscle |
| chr9_+_108368032 | 2.93 |
ENSMUST00000166103.9
ENSMUST00000085044.14 ENSMUST00000193678.6 ENSMUST00000178075.8 |
Usp19
|
ubiquitin specific peptidase 19 |
| chr6_+_70703409 | 2.92 |
ENSMUST00000103410.3
|
Igkc
|
immunoglobulin kappa constant |
| chr9_+_45311000 | 2.91 |
ENSMUST00000216289.2
|
Fxyd2
|
FXYD domain-containing ion transport regulator 2 |
| chr19_+_6450641 | 2.82 |
ENSMUST00000113467.2
|
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chrX_+_156482116 | 2.79 |
ENSMUST00000112521.8
|
Smpx
|
small muscle protein, X-linked |
| chr17_+_8454862 | 2.75 |
ENSMUST00000231340.2
|
Ccr6
|
chemokine (C-C motif) receptor 6 |
| chr1_-_79836344 | 2.74 |
ENSMUST00000027467.11
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chr4_-_131802606 | 2.71 |
ENSMUST00000146021.8
|
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr19_+_6449887 | 2.67 |
ENSMUST00000146601.8
ENSMUST00000150713.8 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr11_-_100713348 | 2.64 |
ENSMUST00000107358.9
|
Stat5b
|
signal transducer and activator of transcription 5B |
| chr9_+_96140781 | 2.62 |
ENSMUST00000190104.7
ENSMUST00000179416.8 ENSMUST00000189606.7 |
Tfdp2
|
transcription factor Dp 2 |
| chrX_-_104918911 | 2.61 |
ENSMUST00000200471.2
|
Atrx
|
ATRX, chromatin remodeler |
| chr5_+_137517140 | 2.59 |
ENSMUST00000031727.10
|
Gigyf1
|
GRB10 interacting GYF protein 1 |
| chr6_+_113508636 | 2.55 |
ENSMUST00000036340.12
ENSMUST00000204827.3 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr17_-_33937565 | 2.51 |
ENSMUST00000174040.2
ENSMUST00000173015.8 ENSMUST00000066121.13 ENSMUST00000186022.7 ENSMUST00000173329.8 ENSMUST00000172767.9 |
Marchf2
|
membrane associated ring-CH-type finger 2 |
| chr9_+_65494469 | 2.47 |
ENSMUST00000239405.2
ENSMUST00000047099.13 ENSMUST00000131483.3 ENSMUST00000141046.3 |
Pif1
|
PIF1 5'-to-3' DNA helicase |
| chr11_-_115503316 | 2.44 |
ENSMUST00000106507.9
|
Mif4gd
|
MIF4G domain containing |
| chr11_+_97340962 | 2.42 |
ENSMUST00000107601.8
|
Arhgap23
|
Rho GTPase activating protein 23 |
| chr1_+_63216281 | 2.39 |
ENSMUST00000188524.2
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr17_-_36220518 | 2.38 |
ENSMUST00000141132.2
|
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr2_+_158344532 | 2.36 |
ENSMUST00000059889.4
|
Adig
|
adipogenin |
| chr17_+_48047955 | 2.36 |
ENSMUST00000086932.10
|
Tfeb
|
transcription factor EB |
| chr1_+_87731360 | 2.36 |
ENSMUST00000177757.2
ENSMUST00000077772.12 |
Sag
|
S-antigen, retina and pineal gland (arrestin) |
| chr11_-_100595019 | 2.35 |
ENSMUST00000017974.13
|
Dhx58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr8_-_105350898 | 2.34 |
ENSMUST00000212882.2
ENSMUST00000163783.4 |
Cdh16
|
cadherin 16 |
| chr19_-_7218363 | 2.27 |
ENSMUST00000236769.2
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr9_+_108367801 | 2.26 |
ENSMUST00000006854.13
|
Usp19
|
ubiquitin specific peptidase 19 |
| chr11_+_70548622 | 2.24 |
ENSMUST00000170716.8
|
Eno3
|
enolase 3, beta muscle |
| chr19_+_6449776 | 2.23 |
ENSMUST00000113468.8
|
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr11_+_68979332 | 2.22 |
ENSMUST00000117780.2
|
Vamp2
|
vesicle-associated membrane protein 2 |
| chr9_-_107474221 | 2.21 |
ENSMUST00000238519.2
|
Lsmem2
|
leucine-rich single-pass membrane protein 2 |
| chr4_-_156340276 | 2.20 |
ENSMUST00000220228.2
ENSMUST00000218788.2 ENSMUST00000179919.3 |
Samd11
|
sterile alpha motif domain containing 11 |
| chr15_-_74624811 | 2.14 |
ENSMUST00000189128.2
ENSMUST00000023259.15 |
Lynx1
|
Ly6/neurotoxin 1 |
| chr17_-_36220924 | 2.14 |
ENSMUST00000141662.8
ENSMUST00000056034.13 ENSMUST00000077494.13 ENSMUST00000149277.8 ENSMUST00000061052.12 |
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr7_-_133833854 | 2.13 |
ENSMUST00000127524.2
|
Adam12
|
a disintegrin and metallopeptidase domain 12 (meltrin alpha) |
| chr11_+_74721733 | 2.13 |
ENSMUST00000000291.9
|
Mnt
|
max binding protein |
| chr10_+_67021509 | 2.10 |
ENSMUST00000173689.8
|
Jmjd1c
|
jumonji domain containing 1C |
| chr11_-_100332619 | 2.09 |
ENSMUST00000107398.8
|
Nt5c3b
|
5'-nucleotidase, cytosolic IIIB |
| chr16_-_20245071 | 2.08 |
ENSMUST00000115547.9
ENSMUST00000096199.5 |
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr1_+_63215976 | 2.07 |
ENSMUST00000129339.8
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr7_-_133833734 | 2.04 |
ENSMUST00000134504.8
|
Adam12
|
a disintegrin and metallopeptidase domain 12 (meltrin alpha) |
| chr19_-_5738177 | 2.04 |
ENSMUST00000068169.12
|
Pcnx3
|
pecanex homolog 3 |
| chr10_+_79984097 | 2.00 |
ENSMUST00000099492.10
ENSMUST00000042057.12 |
Midn
|
midnolin |
| chr10_+_78410803 | 1.97 |
ENSMUST00000218763.2
ENSMUST00000220430.2 ENSMUST00000218885.2 ENSMUST00000218215.2 ENSMUST00000218271.2 |
Ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr1_-_13061333 | 1.96 |
ENSMUST00000115403.9
ENSMUST00000136197.8 ENSMUST00000115402.8 |
Slco5a1
|
solute carrier organic anion transporter family, member 5A1 |
| chr19_+_37184927 | 1.94 |
ENSMUST00000024078.15
ENSMUST00000112391.8 |
Marchf5
|
membrane associated ring-CH-type finger 5 |
| chr2_-_131001916 | 1.94 |
ENSMUST00000103188.10
ENSMUST00000133602.8 ENSMUST00000028800.12 |
1700037H04Rik
|
RIKEN cDNA 1700037H04 gene |
| chr2_+_158636727 | 1.92 |
ENSMUST00000029186.14
ENSMUST00000109478.9 ENSMUST00000156893.2 |
Dhx35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr11_-_48883053 | 1.92 |
ENSMUST00000059930.3
ENSMUST00000068063.4 |
Gm12185
Tgtp1
|
predicted gene 12185 T cell specific GTPase 1 |
| chr14_+_54448904 | 1.92 |
ENSMUST00000103733.2
|
Traj7
|
T cell receptor alpha joining 7 |
| chr3_+_89979948 | 1.91 |
ENSMUST00000121503.8
ENSMUST00000119570.8 |
Tpm3
|
tropomyosin 3, gamma |
| chr7_+_125871761 | 1.90 |
ENSMUST00000056028.11
|
Sbk1
|
SH3-binding kinase 1 |
| chr6_-_39397334 | 1.85 |
ENSMUST00000031985.13
|
Mkrn1
|
makorin, ring finger protein, 1 |
| chr4_-_126096551 | 1.84 |
ENSMUST00000080919.12
|
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr8_+_72021510 | 1.84 |
ENSMUST00000212889.2
|
Slc27a1
|
solute carrier family 27 (fatty acid transporter), member 1 |
| chr8_+_72021567 | 1.82 |
ENSMUST00000034267.5
|
Slc27a1
|
solute carrier family 27 (fatty acid transporter), member 1 |
| chr3_-_129834788 | 1.82 |
ENSMUST00000168644.3
|
Sec24b
|
Sec24 related gene family, member B (S. cerevisiae) |
| chr7_+_28050077 | 1.82 |
ENSMUST00000082134.6
|
Rps16
|
ribosomal protein S16 |
| chrX_+_85235370 | 1.80 |
ENSMUST00000026036.5
|
Nr0b1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr7_-_92319096 | 1.79 |
ENSMUST00000208255.2
ENSMUST00000208058.2 |
Pcf11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr4_-_82939330 | 1.75 |
ENSMUST00000071708.12
|
Frem1
|
Fras1 related extracellular matrix protein 1 |
| chr7_-_92319126 | 1.73 |
ENSMUST00000119954.9
|
Pcf11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr6_-_76474767 | 1.72 |
ENSMUST00000097218.7
|
Gm9008
|
predicted pseudogene 9008 |
| chr15_-_5273659 | 1.72 |
ENSMUST00000047379.15
|
Ptger4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr15_-_5273645 | 1.71 |
ENSMUST00000120563.2
|
Ptger4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr16_-_95387444 | 1.70 |
ENSMUST00000233269.2
|
Erg
|
ETS transcription factor |
| chr4_+_57821050 | 1.70 |
ENSMUST00000238994.2
|
Pakap
|
paralemmin A kinase anchor protein |
| chr9_+_96140750 | 1.70 |
ENSMUST00000186609.7
|
Tfdp2
|
transcription factor Dp 2 |
| chr11_-_114892706 | 1.68 |
ENSMUST00000092464.10
|
Cd300c2
|
CD300C molecule 2 |
| chr6_+_15727798 | 1.67 |
ENSMUST00000128849.3
|
Mdfic
|
MyoD family inhibitor domain containing |
| chrX_+_159551009 | 1.66 |
ENSMUST00000033650.14
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr6_-_39396691 | 1.66 |
ENSMUST00000146785.8
ENSMUST00000114823.8 |
Mkrn1
|
makorin, ring finger protein, 1 |
| chr9_+_75348800 | 1.66 |
ENSMUST00000048937.6
|
Leo1
|
Leo1, Paf1/RNA polymerase II complex component |
| chr3_-_108797306 | 1.64 |
ENSMUST00000102620.10
|
Fndc7
|
fibronectin type III domain containing 7 |
| chr6_+_129374260 | 1.63 |
ENSMUST00000032262.14
|
Clec1b
|
C-type lectin domain family 1, member b |
| chrX_+_100492684 | 1.60 |
ENSMUST00000033674.6
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr18_-_38417390 | 1.60 |
ENSMUST00000025311.8
|
Pcdh12
|
protocadherin 12 |
| chr9_-_110886306 | 1.59 |
ENSMUST00000195968.2
ENSMUST00000111888.3 |
Ccrl2
|
chemokine (C-C motif) receptor-like 2 |
| chr9_-_58277734 | 1.58 |
ENSMUST00000040217.6
|
Tbc1d21
|
TBC1 domain family, member 21 |
| chr4_-_116228921 | 1.57 |
ENSMUST00000239239.2
ENSMUST00000239177.2 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr5_+_37242714 | 1.54 |
ENSMUST00000121010.9
ENSMUST00000174629.2 ENSMUST00000232332.3 |
Jakmip1
Gm1043
|
janus kinase and microtubule interacting protein 1 predicted gene 1043 |
| chr6_-_39397212 | 1.54 |
ENSMUST00000114822.2
ENSMUST00000051671.11 |
Mkrn1
|
makorin, ring finger protein, 1 |
| chr11_+_70548513 | 1.54 |
ENSMUST00000134087.8
|
Eno3
|
enolase 3, beta muscle |
| chr6_+_129374441 | 1.54 |
ENSMUST00000112081.9
ENSMUST00000112079.3 |
Clec1b
|
C-type lectin domain family 1, member b |
| chr5_-_113163339 | 1.48 |
ENSMUST00000197776.2
ENSMUST00000065167.9 |
Grk3
|
G protein-coupled receptor kinase 3 |
| chr15_+_79578141 | 1.47 |
ENSMUST00000230898.2
ENSMUST00000229046.2 |
Gtpbp1
|
GTP binding protein 1 |
| chr9_+_108270020 | 1.47 |
ENSMUST00000035234.6
|
1700102P08Rik
|
RIKEN cDNA 1700102P08 gene |
| chr4_+_33310306 | 1.47 |
ENSMUST00000108153.9
ENSMUST00000029942.8 |
Rngtt
|
RNA guanylyltransferase and 5'-phosphatase |
| chr9_+_109891628 | 1.46 |
ENSMUST00000163979.7
|
Map4
|
microtubule-associated protein 4 |
| chr10_+_126911134 | 1.45 |
ENSMUST00000239120.2
|
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr17_-_56891576 | 1.45 |
ENSMUST00000075510.12
|
Safb2
|
scaffold attachment factor B2 |
| chr10_-_12745109 | 1.44 |
ENSMUST00000218635.2
|
Utrn
|
utrophin |
| chr9_+_65494429 | 1.40 |
ENSMUST00000134538.9
|
Pif1
|
PIF1 5'-to-3' DNA helicase |
| chr12_+_80739365 | 1.40 |
ENSMUST00000140770.2
|
Plekhd1
|
pleckstrin homology domain containing, family D (with coiled-coil domains) member 1 |
| chr15_-_50746202 | 1.39 |
ENSMUST00000184885.8
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chrX_+_108240356 | 1.39 |
ENSMUST00000139259.2
ENSMUST00000060013.4 |
Gm6377
|
predicted gene 6377 |
| chr4_-_126096376 | 1.38 |
ENSMUST00000106142.8
ENSMUST00000169403.8 ENSMUST00000130334.2 |
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr10_-_128383508 | 1.37 |
ENSMUST00000152539.8
ENSMUST00000133458.2 ENSMUST00000040572.10 |
Zc3h10
|
zinc finger CCCH type containing 10 |
| chr18_-_65527078 | 1.36 |
ENSMUST00000035548.16
|
Alpk2
|
alpha-kinase 2 |
| chr7_-_140676623 | 1.36 |
ENSMUST00000209352.2
|
Sigirr
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr2_+_132532040 | 1.35 |
ENSMUST00000148271.8
ENSMUST00000110132.3 |
Shld1
|
shieldin complex subunit 1 |
| chr2_+_158344553 | 1.35 |
ENSMUST00000109484.2
|
Adig
|
adipogenin |
| chr11_+_70655035 | 1.27 |
ENSMUST00000060444.6
|
Zfp3
|
zinc finger protein 3 |
| chr2_+_130248398 | 1.26 |
ENSMUST00000055421.6
|
Tmem239
|
transmembrane 239 |
| chr1_-_131025562 | 1.26 |
ENSMUST00000016672.11
|
Mapkapk2
|
MAP kinase-activated protein kinase 2 |
| chr11_-_94568228 | 1.25 |
ENSMUST00000116349.9
|
Xylt2
|
xylosyltransferase II |
| chr10_-_93727003 | 1.25 |
ENSMUST00000180840.8
|
Metap2
|
methionine aminopeptidase 2 |
| chr5_+_33021042 | 1.25 |
ENSMUST00000149350.8
ENSMUST00000118698.8 ENSMUST00000150130.8 ENSMUST00000049780.13 ENSMUST00000087897.11 ENSMUST00000119705.8 ENSMUST00000125574.8 |
Depdc5
|
DEP domain containing 5 |
| chr11_-_115503739 | 1.23 |
ENSMUST00000021087.14
|
Mif4gd
|
MIF4G domain containing |
| chr9_-_22042930 | 1.23 |
ENSMUST00000213815.2
|
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr2_+_160573604 | 1.23 |
ENSMUST00000174885.2
ENSMUST00000109462.8 |
Plcg1
|
phospholipase C, gamma 1 |
| chr16_+_43960183 | 1.23 |
ENSMUST00000159514.8
ENSMUST00000161326.8 ENSMUST00000063520.15 ENSMUST00000063542.8 |
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr9_-_22043083 | 1.22 |
ENSMUST00000069330.14
ENSMUST00000217643.2 |
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr17_+_48769383 | 1.22 |
ENSMUST00000162132.8
|
Unc5cl
|
unc-5 family C-terminal like |
| chr18_-_38417444 | 1.21 |
ENSMUST00000194012.2
|
Pcdh12
|
protocadherin 12 |
| chr3_+_90507510 | 1.21 |
ENSMUST00000001047.8
|
S100a3
|
S100 calcium binding protein A3 |
| chr10_+_11157047 | 1.19 |
ENSMUST00000129456.8
|
Fbxo30
|
F-box protein 30 |
| chrX_+_159551171 | 1.17 |
ENSMUST00000112368.3
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr1_+_93096316 | 1.16 |
ENSMUST00000138595.3
|
Crocc2
|
ciliary rootlet coiled-coil, rootletin family member 2 |
| chr15_-_63932176 | 1.16 |
ENSMUST00000226675.2
ENSMUST00000228226.2 ENSMUST00000227024.2 |
Cyrib
|
CYFIP related Rac1 interactor B |
| chrX_-_36368176 | 1.16 |
ENSMUST00000130324.2
|
Upf3b
|
UPF3 regulator of nonsense transcripts homolog B (yeast) |
| chr7_-_134100659 | 1.15 |
ENSMUST00000238294.2
|
D7Ertd443e
|
DNA segment, Chr 7, ERATO Doi 443, expressed |
| chr14_-_70867588 | 1.12 |
ENSMUST00000228009.2
|
Dmtn
|
dematin actin binding protein |
| chr3_-_32791296 | 1.12 |
ENSMUST00000043966.8
|
Mrpl47
|
mitochondrial ribosomal protein L47 |
| chr2_+_61423421 | 1.11 |
ENSMUST00000112495.8
ENSMUST00000112501.9 |
Tank
|
TRAF family member-associated Nf-kappa B activator |
| chr4_-_128856213 | 1.10 |
ENSMUST00000119354.8
ENSMUST00000106068.8 ENSMUST00000030581.10 |
Azin2
|
antizyme inhibitor 2 |
| chr16_+_27208660 | 1.10 |
ENSMUST00000143823.2
|
Ccdc50
|
coiled-coil domain containing 50 |
| chr11_+_100332709 | 1.09 |
ENSMUST00000001599.4
ENSMUST00000107395.3 |
Klhl10
|
kelch-like 10 |
| chr11_+_94218810 | 1.09 |
ENSMUST00000107818.9
ENSMUST00000051221.13 |
Ankrd40
|
ankyrin repeat domain 40 |
| chr2_+_79538124 | 1.09 |
ENSMUST00000090760.9
ENSMUST00000040863.11 ENSMUST00000111780.3 |
Ppp1r1c
|
protein phosphatase 1, regulatory inhibitor subunit 1C |
| chr19_-_37184692 | 1.03 |
ENSMUST00000132580.8
ENSMUST00000079754.11 ENSMUST00000136286.8 ENSMUST00000126188.8 ENSMUST00000126781.2 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr15_+_99600149 | 1.02 |
ENSMUST00000229236.2
|
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr6_-_39396902 | 1.01 |
ENSMUST00000122996.8
|
Mkrn1
|
makorin, ring finger protein, 1 |
| chr14_+_54393590 | 0.98 |
ENSMUST00000197589.2
ENSMUST00000103686.2 |
Traj59
|
T cell receptor alpha joining 59 |
| chr4_-_138546564 | 0.96 |
ENSMUST00000102512.11
|
Pla2g5
|
phospholipase A2, group V |
| chr17_+_33857030 | 0.96 |
ENSMUST00000052079.8
|
Pram1
|
PML-RAR alpha-regulated adaptor molecule 1 |
| chr11_+_68979308 | 0.95 |
ENSMUST00000021273.13
|
Vamp2
|
vesicle-associated membrane protein 2 |
| chr16_+_17093941 | 0.95 |
ENSMUST00000164950.11
|
Tmem191c
|
transmembrane protein 191C |
| chr8_+_95703728 | 0.94 |
ENSMUST00000179619.9
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr5_-_122917341 | 0.94 |
ENSMUST00000198257.5
ENSMUST00000199599.2 ENSMUST00000196742.2 ENSMUST00000200109.5 ENSMUST00000111668.8 |
Camkk2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta |
| chr8_+_27467330 | 0.93 |
ENSMUST00000127097.9
ENSMUST00000154256.3 |
Zfp703
|
zinc finger protein 703 |
| chr2_-_121637469 | 0.93 |
ENSMUST00000110592.2
|
Frmd5
|
FERM domain containing 5 |
| chr6_-_29164981 | 0.92 |
ENSMUST00000007993.16
|
Rbm28
|
RNA binding motif protein 28 |
| chr8_-_71938598 | 0.88 |
ENSMUST00000093450.6
ENSMUST00000213382.2 |
Ano8
|
anoctamin 8 |
| chr11_+_113550109 | 0.88 |
ENSMUST00000137878.2
|
Cog1
|
component of oligomeric golgi complex 1 |
| chr14_+_54439232 | 0.88 |
ENSMUST00000103724.2
|
Traj17
|
T cell receptor alpha joining 17 |
| chr7_+_139798321 | 0.88 |
ENSMUST00000209637.2
|
Scart1
|
scavenger receptor family member expressed on T cells 1 |
| chrX_+_156481906 | 0.87 |
ENSMUST00000136141.2
ENSMUST00000190091.7 |
Smpx
|
small muscle protein, X-linked |
| chr17_-_25105277 | 0.86 |
ENSMUST00000234583.2
ENSMUST00000234968.2 ENSMUST00000044252.7 |
Nubp2
|
nucleotide binding protein 2 |
| chr15_+_99600475 | 0.85 |
ENSMUST00000228984.2
ENSMUST00000229845.2 |
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr1_+_131838220 | 0.85 |
ENSMUST00000189946.7
|
Nucks1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr14_-_70680882 | 0.83 |
ENSMUST00000000793.13
|
Polr3d
|
polymerase (RNA) III (DNA directed) polypeptide D |
| chr8_+_82582953 | 0.83 |
ENSMUST00000109851.3
|
Inpp4b
|
inositol polyphosphate-4-phosphatase, type II |
| chr3_+_36917251 | 0.83 |
ENSMUST00000057272.15
|
4932438A13Rik
|
RIKEN cDNA 4932438A13 gene |
| chr15_+_99122742 | 0.83 |
ENSMUST00000041415.5
|
Kcnh3
|
potassium voltage-gated channel, subfamily H (eag-related), member 3 |
| chr18_+_36877709 | 0.82 |
ENSMUST00000007042.6
ENSMUST00000237095.2 |
Ik
|
IK cytokine |
| chr5_+_115644727 | 0.81 |
ENSMUST00000067268.15
ENSMUST00000086523.7 ENSMUST00000212819.3 |
Pxn
|
paxillin |
| chr2_-_121637505 | 0.81 |
ENSMUST00000138157.8
|
Frmd5
|
FERM domain containing 5 |
| chr6_+_67993691 | 0.81 |
ENSMUST00000103314.3
|
Igkv1-122
|
immunoglobulin kappa chain variable 1-122 |
| chr5_+_53747556 | 0.80 |
ENSMUST00000037618.13
ENSMUST00000201912.4 |
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr18_-_43610829 | 0.80 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr11_+_75084609 | 0.80 |
ENSMUST00000102514.4
|
Rtn4rl1
|
reticulon 4 receptor-like 1 |
| chr5_-_121665249 | 0.79 |
ENSMUST00000152270.8
|
Mapkapk5
|
MAP kinase-activated protein kinase 5 |
| chr4_-_156340713 | 0.79 |
ENSMUST00000219393.2
|
Samd11
|
sterile alpha motif domain containing 11 |
| chr9_-_54568950 | 0.78 |
ENSMUST00000128624.2
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr14_+_54423447 | 0.77 |
ENSMUST00000103709.2
|
Traj32
|
T cell receptor alpha joining 32 |
| chr13_-_64645606 | 0.77 |
ENSMUST00000180282.2
|
Fam240b
|
family with sequence similarity 240 member B |
| chr16_-_10609959 | 0.77 |
ENSMUST00000037996.7
|
Prm2
|
protamine 2 |
| chr18_-_67857594 | 0.76 |
ENSMUST00000120934.8
ENSMUST00000025420.14 ENSMUST00000122412.2 |
Ptpn2
|
protein tyrosine phosphatase, non-receptor type 2 |
| chr9_-_110886576 | 0.75 |
ENSMUST00000199839.5
|
Ccrl2
|
chemokine (C-C motif) receptor-like 2 |
| chr2_+_130418091 | 0.75 |
ENSMUST00000028764.6
|
Oxt
|
oxytocin |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.9 | GO:0071846 | actin filament debranching(GO:0071846) |
| 2.3 | 9.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 2.2 | 6.7 | GO:1904156 | DN2 thymocyte differentiation(GO:1904155) DN3 thymocyte differentiation(GO:1904156) |
| 1.5 | 4.5 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 1.1 | 3.4 | GO:2000387 | negative regulation of integrin activation(GO:0033624) regulation of matrix metallopeptidase secretion(GO:1904464) matrix metallopeptidase secretion(GO:1990773) positive regulation of ovarian follicle development(GO:2000386) regulation of antral ovarian follicle growth(GO:2000387) positive regulation of antral ovarian follicle growth(GO:2000388) negative regulation of eosinophil migration(GO:2000417) |
| 1.0 | 4.0 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.9 | 5.6 | GO:0035128 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.9 | 3.7 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.9 | 2.7 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.8 | 7.8 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.8 | 0.8 | GO:0070104 | negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.7 | 5.0 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.7 | 2.8 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.7 | 5.9 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.6 | 2.4 | GO:0009816 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.6 | 6.8 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.5 | 3.1 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.5 | 3.3 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.5 | 1.8 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.5 | 1.8 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.5 | 3.2 | GO:0002278 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil activation(GO:0043307) eosinophil degranulation(GO:0043308) |
| 0.4 | 3.4 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.4 | 2.4 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.4 | 3.1 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.4 | 3.9 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.4 | 6.1 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.3 | 3.5 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.3 | 0.9 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.3 | 2.4 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.3 | 1.5 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.3 | 1.7 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.3 | 1.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.3 | 2.5 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.3 | 0.8 | GO:0060450 | positive regulation of hindgut contraction(GO:0060450) |
| 0.2 | 2.0 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.2 | 1.2 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.2 | 5.2 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.2 | 0.7 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.2 | 0.8 | GO:0019046 | release from viral latency(GO:0019046) regulation of DNA strand elongation(GO:0060382) |
| 0.2 | 1.5 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 1.3 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.2 | 1.4 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.2 | 1.8 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.2 | 2.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.2 | 1.1 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.2 | 1.1 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.2 | 0.2 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.2 | 0.5 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.2 | 0.7 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.2 | 0.5 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.2 | 1.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.2 | 0.8 | GO:1905068 | positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.2 | 2.7 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 3.3 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.1 | 1.5 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 0.8 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 2.4 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.1 | 3.5 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.1 | 4.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 3.7 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.1 | 0.7 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.6 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 0.8 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.1 | 0.7 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 1.3 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 3.2 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 0.3 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.1 | 1.6 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 | 0.3 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.1 | 1.9 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 0.7 | GO:0046103 | inosine biosynthetic process(GO:0046103) |
| 0.1 | 1.7 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.1 | 3.2 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 3.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 7.2 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 1.2 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.1 | 1.0 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) regulation of neutrophil activation(GO:1902563) |
| 0.1 | 1.7 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.1 | 5.3 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 0.1 | 1.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.7 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 1.8 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 1.9 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.1 | 0.7 | GO:0002827 | positive regulation of T-helper 1 type immune response(GO:0002827) |
| 0.1 | 2.1 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 0.6 | GO:0045625 | regulation of T-helper 1 cell differentiation(GO:0045625) |
| 0.1 | 6.2 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 7.2 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.1 | 0.2 | GO:0061193 | taste bud development(GO:0061193) |
| 0.1 | 1.1 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 2.6 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.1 | 3.4 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 0.4 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.1 | 0.8 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 1.7 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.3 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.3 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.8 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 1.0 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 1.8 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.8 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.9 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 1.5 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.8 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 1.2 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 6.6 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.6 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 2.1 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 1.1 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.3 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 2.9 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.5 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:2000983 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.0 | 0.9 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.8 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 1.7 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 1.6 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 1.2 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.0 | 0.8 | GO:0051647 | nucleus localization(GO:0051647) |
| 0.0 | 0.1 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.4 | GO:0097499 | protein localization to nonmotile primary cilium(GO:0097499) |
| 0.0 | 0.9 | GO:0051384 | response to glucocorticoid(GO:0051384) |
| 0.0 | 3.7 | GO:0006417 | regulation of translation(GO:0006417) |
| 0.0 | 0.5 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:1990879 | CST complex(GO:1990879) |
| 1.1 | 7.4 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.9 | 5.6 | GO:1990707 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.6 | 4.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.6 | 3.2 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.6 | 7.5 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.5 | 6.8 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.5 | 3.7 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.3 | 4.5 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.3 | 3.4 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.3 | 0.8 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.3 | 1.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 0.7 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.2 | 6.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 2.7 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.2 | 1.7 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.2 | 1.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 7.8 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 0.7 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 1.9 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 1.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 1.8 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 3.5 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.1 | 4.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 2.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 5.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 1.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 1.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.8 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 1.0 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.1 | 0.8 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 0.9 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 1.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 1.5 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.7 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 3.9 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.3 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 6.7 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 5.5 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.8 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 3.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.6 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 1.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 7.9 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 3.7 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 2.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 1.4 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.6 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 5.4 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 2.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 2.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 2.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.7 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 4.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0033680 | ATP-dependent DNA/RNA helicase activity(GO:0033680) |
| 1.1 | 7.4 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.8 | 5.6 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.8 | 10.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.7 | 3.0 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.5 | 3.4 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.5 | 1.5 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.5 | 6.8 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.5 | 6.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.5 | 2.4 | GO:0002046 | opsin binding(GO:0002046) |
| 0.5 | 7.8 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.5 | 1.8 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.4 | 1.3 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.4 | 4.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.4 | 5.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.3 | 5.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.3 | 1.5 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.3 | 1.7 | GO:1990269 | RNA polymerase II C-terminal domain phosphoserine binding(GO:1990269) |
| 0.3 | 1.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.3 | 3.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.3 | 2.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.3 | 2.8 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.3 | 0.8 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.2 | 3.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 2.3 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.2 | 2.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 0.8 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.2 | 0.8 | GO:0051435 | BH4 domain binding(GO:0051435) |
| 0.2 | 0.7 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.2 | 4.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 2.0 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 0.7 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 0.8 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 2.4 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 1.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 2.0 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.8 | GO:0002135 | CTP binding(GO:0002135) |
| 0.1 | 0.3 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.1 | 1.2 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 3.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 1.0 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 1.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 3.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 2.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 3.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 2.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 2.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 5.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 1.7 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 1.5 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.1 | 0.7 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 2.9 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.8 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 1.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 0.2 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 0.1 | 2.1 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 1.8 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.1 | 7.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 1.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.6 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 5.9 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 6.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.8 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 4.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 8.4 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.0 | 0.9 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 5.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 1.1 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.5 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 1.9 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 1.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.3 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.0 | 2.5 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 4.1 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 1.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 2.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 6.6 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.6 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.9 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 2.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.1 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.2 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 11.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 5.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 9.5 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 10.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 5.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 3.3 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.1 | 3.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 2.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 2.7 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 2.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 2.5 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 9.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 0.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 1.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 11.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 4.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 3.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 6.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.8 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.6 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 6.7 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.3 | 3.4 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.3 | 11.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.3 | 5.8 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.3 | 1.3 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.3 | 7.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 4.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.2 | 3.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.2 | 2.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.2 | 4.6 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.2 | 6.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 7.8 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 9.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 6.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.8 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 0.8 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 1.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 3.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 5.8 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 0.8 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 1.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 1.0 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 1.5 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.1 | 3.5 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.0 | 0.8 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.8 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.7 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 2.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 6.3 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 6.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 2.7 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 1.2 | REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | Genes involved in MyD88:Mal cascade initiated on plasma membrane |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |