Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Rorc_Nr1d1
Z-value: 0.79
Transcription factors associated with Rorc_Nr1d1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
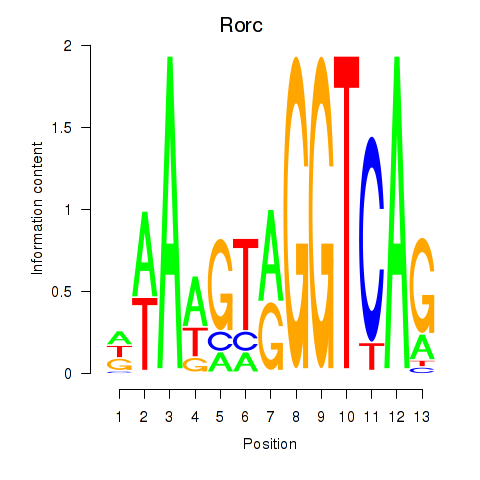
Rorc
|
ENSMUSG00000028150.15 | Rorc |
|
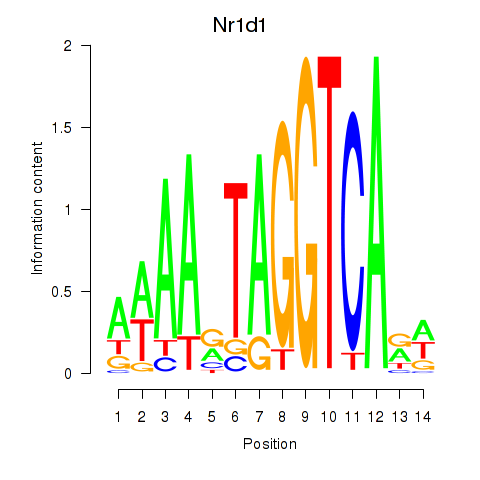
Nr1d1
|
ENSMUSG00000020889.12 | Nr1d1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr1d1 | mm39_v1_chr11_-_98666159_98666183 | 0.16 | 1.8e-01 | Click! |
| Rorc | mm39_v1_chr3_+_94280101_94280180 | 0.06 | 6.0e-01 | Click! |
Activity profile of Rorc_Nr1d1 motif
Sorted Z-values of Rorc_Nr1d1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Rorc_Nr1d1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_105249308 | 7.57 |
ENSMUST00000210531.2
ENSMUST00000033185.10 |
Hpx
|
hemopexin |
| chr7_-_28824440 | 7.23 |
ENSMUST00000214374.2
ENSMUST00000179893.9 ENSMUST00000032813.10 |
Ryr1
|
ryanodine receptor 1, skeletal muscle |
| chr5_+_90708962 | 3.93 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr11_+_4833186 | 3.65 |
ENSMUST00000139737.2
|
Nipsnap1
|
nipsnap homolog 1 |
| chrX_-_135104589 | 3.37 |
ENSMUST00000066819.11
|
Tceal5
|
transcription elongation factor A (SII)-like 5 |
| chrX_-_135104386 | 3.13 |
ENSMUST00000151592.8
ENSMUST00000131510.2 |
Tceal5
|
transcription elongation factor A (SII)-like 5 |
| chr7_+_126808016 | 3.09 |
ENSMUST00000206204.2
ENSMUST00000206772.2 |
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr1_-_14380418 | 3.09 |
ENSMUST00000027066.13
ENSMUST00000168081.9 |
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr7_+_126810780 | 2.97 |
ENSMUST00000032910.13
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr7_+_45354512 | 2.91 |
ENSMUST00000080885.12
ENSMUST00000211513.2 ENSMUST00000211357.2 |
Dbp
|
D site albumin promoter binding protein |
| chr19_-_40175709 | 2.72 |
ENSMUST00000051846.13
|
Cyp2c70
|
cytochrome P450, family 2, subfamily c, polypeptide 70 |
| chr7_-_99276310 | 2.72 |
ENSMUST00000178124.3
|
Tpbgl
|
trophoblast glycoprotein-like |
| chr6_+_71176811 | 2.65 |
ENSMUST00000067492.8
|
Fabp1
|
fatty acid binding protein 1, liver |
| chr10_+_106306122 | 2.54 |
ENSMUST00000029404.17
ENSMUST00000217854.2 |
Ppfia2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr1_-_14380327 | 2.46 |
ENSMUST00000080664.14
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr16_-_20549294 | 2.38 |
ENSMUST00000231826.2
ENSMUST00000076422.13 ENSMUST00000232217.2 |
Thpo
|
thrombopoietin |
| chr11_-_4045343 | 2.35 |
ENSMUST00000004868.6
|
Mtfp1
|
mitochondrial fission process 1 |
| chr5_+_31454787 | 2.29 |
ENSMUST00000201166.4
ENSMUST00000072228.9 |
Gckr
|
glucokinase regulatory protein |
| chr17_-_56440817 | 2.07 |
ENSMUST00000167545.3
|
Sema6b
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
| chr2_+_158344553 | 2.04 |
ENSMUST00000109484.2
|
Adig
|
adipogenin |
| chr19_+_6434416 | 1.87 |
ENSMUST00000035269.15
ENSMUST00000113483.2 |
Pygm
|
muscle glycogen phosphorylase |
| chr13_-_63036096 | 1.76 |
ENSMUST00000092888.11
|
Fbp1
|
fructose bisphosphatase 1 |
| chr1_-_180021039 | 1.75 |
ENSMUST00000160482.8
ENSMUST00000170472.8 |
Coq8a
|
coenzyme Q8A |
| chrX_-_16777913 | 1.73 |
ENSMUST00000040134.8
|
Ndp
|
Norrie disease (pseudoglioma) (human) |
| chr3_-_72965136 | 1.68 |
ENSMUST00000059407.9
|
Slitrk3
|
SLIT and NTRK-like family, member 3 |
| chr1_-_180021218 | 1.64 |
ENSMUST00000159914.8
|
Coq8a
|
coenzyme Q8A |
| chr13_+_83720484 | 1.59 |
ENSMUST00000196207.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr2_+_96148418 | 1.58 |
ENSMUST00000135431.8
ENSMUST00000162807.9 |
Lrrc4c
|
leucine rich repeat containing 4C |
| chr2_+_90716204 | 1.57 |
ENSMUST00000111466.3
|
C1qtnf4
|
C1q and tumor necrosis factor related protein 4 |
| chr11_-_115518774 | 1.55 |
ENSMUST00000154623.2
ENSMUST00000106503.10 ENSMUST00000141614.3 |
Slc25a19
|
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
| chr14_-_51134930 | 1.50 |
ENSMUST00000227271.2
|
Klhl33
|
kelch-like 33 |
| chr11_-_98220466 | 1.50 |
ENSMUST00000041685.7
|
Neurod2
|
neurogenic differentiation 2 |
| chr6_-_88851027 | 1.44 |
ENSMUST00000038409.12
|
Podxl2
|
podocalyxin-like 2 |
| chr18_-_12995261 | 1.43 |
ENSMUST00000234427.2
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr1_-_14380032 | 1.43 |
ENSMUST00000187790.2
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr13_-_21257469 | 1.39 |
ENSMUST00000058168.2
|
Olfr1370
|
olfactory receptor 1370 |
| chr1_-_162305573 | 1.39 |
ENSMUST00000086074.12
ENSMUST00000070330.14 |
Dnm3
|
dynamin 3 |
| chr4_+_115641996 | 1.37 |
ENSMUST00000177280.8
ENSMUST00000176047.8 |
Atpaf1
|
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr7_+_114367971 | 1.33 |
ENSMUST00000117543.3
ENSMUST00000151464.2 |
Insc
|
INSC spindle orientation adaptor protein |
| chr11_-_115518951 | 1.31 |
ENSMUST00000155709.2
ENSMUST00000021089.11 |
Slc25a19
|
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
| chr2_-_5680801 | 1.31 |
ENSMUST00000114987.4
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr3_-_107838895 | 1.30 |
ENSMUST00000133947.9
ENSMUST00000124215.2 ENSMUST00000106688.8 ENSMUST00000106687.9 |
Gstm7
|
glutathione S-transferase, mu 7 |
| chr17_-_46342739 | 1.30 |
ENSMUST00000024747.14
|
Vegfa
|
vascular endothelial growth factor A |
| chr7_-_142215027 | 1.29 |
ENSMUST00000105936.8
|
Igf2
|
insulin-like growth factor 2 |
| chr2_+_158344532 | 1.27 |
ENSMUST00000059889.4
|
Adig
|
adipogenin |
| chr11_-_115519086 | 1.24 |
ENSMUST00000178003.8
|
Slc25a19
|
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
| chr1_-_120432669 | 1.22 |
ENSMUST00000027639.8
|
Marco
|
macrophage receptor with collagenous structure |
| chr9_-_100916910 | 1.19 |
ENSMUST00000142676.8
ENSMUST00000149322.8 ENSMUST00000189498.2 |
Pccb
|
propionyl Coenzyme A carboxylase, beta polypeptide |
| chr5_+_125608838 | 1.16 |
ENSMUST00000185104.2
|
Tmem132b
|
transmembrane protein 132B |
| chr16_+_39804711 | 1.15 |
ENSMUST00000187695.7
|
Lsamp
|
limbic system-associated membrane protein |
| chr13_+_6598185 | 1.15 |
ENSMUST00000021611.10
ENSMUST00000222485.2 |
Pitrm1
|
pitrilysin metallepetidase 1 |
| chr11_-_119907884 | 1.11 |
ENSMUST00000132575.8
|
Aatk
|
apoptosis-associated tyrosine kinase |
| chr17_-_46343291 | 1.10 |
ENSMUST00000071648.12
ENSMUST00000142351.9 |
Vegfa
|
vascular endothelial growth factor A |
| chr10_+_73657689 | 1.10 |
ENSMUST00000064562.14
ENSMUST00000193174.6 ENSMUST00000105426.10 ENSMUST00000129404.9 ENSMUST00000131321.9 ENSMUST00000126920.9 ENSMUST00000147189.9 ENSMUST00000105424.10 ENSMUST00000092420.13 ENSMUST00000105429.10 ENSMUST00000193361.6 ENSMUST00000131724.9 ENSMUST00000152655.9 ENSMUST00000151116.9 ENSMUST00000155701.9 ENSMUST00000152819.9 ENSMUST00000125517.9 ENSMUST00000124046.8 ENSMUST00000146682.8 ENSMUST00000177107.8 ENSMUST00000149977.9 ENSMUST00000191854.6 |
Pcdh15
|
protocadherin 15 |
| chrX_-_37665041 | 1.08 |
ENSMUST00000050083.6
ENSMUST00000115118.8 |
Cul4b
|
cullin 4B |
| chr14_+_79086492 | 1.07 |
ENSMUST00000040990.7
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr11_-_116089866 | 1.04 |
ENSMUST00000066587.12
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr9_-_100916946 | 1.02 |
ENSMUST00000035116.12
|
Pccb
|
propionyl Coenzyme A carboxylase, beta polypeptide |
| chr17_-_45997132 | 0.99 |
ENSMUST00000113523.9
|
Tmem63b
|
transmembrane protein 63b |
| chr7_-_133833734 | 0.97 |
ENSMUST00000134504.8
|
Adam12
|
a disintegrin and metallopeptidase domain 12 (meltrin alpha) |
| chr15_-_77290765 | 0.95 |
ENSMUST00000231161.2
|
Apol9a
|
apolipoprotein L 9a |
| chr11_-_116089595 | 0.94 |
ENSMUST00000072948.11
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr13_-_53135064 | 0.93 |
ENSMUST00000071065.8
|
Nfil3
|
nuclear factor, interleukin 3, regulated |
| chr3_-_107839133 | 0.93 |
ENSMUST00000004137.11
|
Gstm7
|
glutathione S-transferase, mu 7 |
| chr14_-_51134906 | 0.90 |
ENSMUST00000170855.2
|
Klhl33
|
kelch-like 33 |
| chr1_-_130815144 | 0.89 |
ENSMUST00000121040.8
|
Il24
|
interleukin 24 |
| chr10_-_116808514 | 0.87 |
ENSMUST00000092165.5
|
Gm10271
|
predicted gene 10271 |
| chr1_-_120432477 | 0.86 |
ENSMUST00000186432.3
|
Marco
|
macrophage receptor with collagenous structure |
| chr14_-_55762416 | 0.85 |
ENSMUST00000178694.3
|
Nrl
|
neural retina leucine zipper gene |
| chr1_+_165123358 | 0.84 |
ENSMUST00000178700.8
|
Gpr161
|
G protein-coupled receptor 161 |
| chr3_+_137573436 | 0.84 |
ENSMUST00000090178.10
|
Dnajb14
|
DnaJ heat shock protein family (Hsp40) member B14 |
| chr11_-_100418688 | 0.84 |
ENSMUST00000107385.2
|
Acly
|
ATP citrate lyase |
| chr14_-_55762432 | 0.82 |
ENSMUST00000062232.15
|
Nrl
|
neural retina leucine zipper gene |
| chr19_+_22425534 | 0.81 |
ENSMUST00000235522.2
ENSMUST00000236372.2 ENSMUST00000238066.2 ENSMUST00000235780.2 ENSMUST00000236804.2 |
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr18_+_35987791 | 0.78 |
ENSMUST00000235404.2
|
Cxxc5
|
CXXC finger 5 |
| chr12_-_72455708 | 0.78 |
ENSMUST00000078505.14
|
Rtn1
|
reticulon 1 |
| chr5_-_122917341 | 0.76 |
ENSMUST00000198257.5
ENSMUST00000199599.2 ENSMUST00000196742.2 ENSMUST00000200109.5 ENSMUST00000111668.8 |
Camkk2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta |
| chr3_-_89300936 | 0.76 |
ENSMUST00000124783.8
ENSMUST00000126027.8 |
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chrX_+_69429475 | 0.76 |
ENSMUST00000053981.6
|
Eola1
|
endothelium and lymphocyte associated ASCH domain 1 |
| chr17_+_47680097 | 0.73 |
ENSMUST00000060752.13
ENSMUST00000119841.8 |
Mrps10
|
mitochondrial ribosomal protein S10 |
| chr4_-_33189390 | 0.72 |
ENSMUST00000098181.9
|
Pm20d2
|
peptidase M20 domain containing 2 |
| chr11_+_69920956 | 0.71 |
ENSMUST00000232115.2
|
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr19_+_31846154 | 0.70 |
ENSMUST00000224564.2
ENSMUST00000224304.2 ENSMUST00000075838.8 ENSMUST00000224400.2 |
A1cf
|
APOBEC1 complementation factor |
| chr18_-_39000056 | 0.69 |
ENSMUST00000236630.2
ENSMUST00000237356.2 |
Fgf1
|
fibroblast growth factor 1 |
| chr1_+_6805048 | 0.68 |
ENSMUST00000139838.8
|
St18
|
suppression of tumorigenicity 18 |
| chrX_-_142716200 | 0.68 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr2_-_167030706 | 0.67 |
ENSMUST00000207917.2
|
Kcnb1
|
potassium voltage gated channel, Shab-related subfamily, member 1 |
| chr19_+_22425565 | 0.66 |
ENSMUST00000037901.14
|
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr15_+_16728842 | 0.65 |
ENSMUST00000228307.2
|
Cdh9
|
cadherin 9 |
| chr16_-_37205277 | 0.63 |
ENSMUST00000114787.8
ENSMUST00000114782.8 ENSMUST00000114775.8 |
Stxbp5l
|
syntaxin binding protein 5-like |
| chr14_-_55762395 | 0.63 |
ENSMUST00000228287.2
|
Nrl
|
neural retina leucine zipper gene |
| chr17_+_25339734 | 0.62 |
ENSMUST00000054930.7
ENSMUST00000232902.2 ENSMUST00000233502.2 |
Ptx4
|
pentraxin 4 |
| chr11_-_100418717 | 0.60 |
ENSMUST00000107389.8
ENSMUST00000007131.16 |
Acly
|
ATP citrate lyase |
| chr16_-_37205302 | 0.60 |
ENSMUST00000114781.8
ENSMUST00000114780.8 |
Stxbp5l
|
syntaxin binding protein 5-like |
| chr3_-_153650068 | 0.59 |
ENSMUST00000150070.2
|
Acadm
|
acyl-Coenzyme A dehydrogenase, medium chain |
| chr11_+_87471867 | 0.58 |
ENSMUST00000018544.12
ENSMUST00000063156.11 ENSMUST00000107960.8 |
Septin4
|
septin 4 |
| chr17_+_23819861 | 0.57 |
ENSMUST00000123866.8
|
Zscan10
|
zinc finger and SCAN domain containing 10 |
| chr7_-_133833854 | 0.57 |
ENSMUST00000127524.2
|
Adam12
|
a disintegrin and metallopeptidase domain 12 (meltrin alpha) |
| chr7_+_28393419 | 0.57 |
ENSMUST00000108280.2
|
Fbxo27
|
F-box protein 27 |
| chrX_-_142716085 | 0.57 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr2_-_160714749 | 0.56 |
ENSMUST00000176141.8
|
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr11_+_69920849 | 0.55 |
ENSMUST00000143920.4
|
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr8_-_85807281 | 0.53 |
ENSMUST00000152785.8
|
Wdr83
|
WD repeat domain containing 83 |
| chr2_+_4394425 | 0.52 |
ENSMUST00000115037.9
|
Frmd4a
|
FERM domain containing 4A |
| chr7_-_142215595 | 0.51 |
ENSMUST00000145896.3
|
Igf2
|
insulin-like growth factor 2 |
| chr9_-_38894338 | 0.51 |
ENSMUST00000074211.2
|
Olfr934
|
olfactory receptor 934 |
| chr5_-_5564873 | 0.50 |
ENSMUST00000060947.14
|
Cldn12
|
claudin 12 |
| chr18_-_65527078 | 0.48 |
ENSMUST00000035548.16
|
Alpk2
|
alpha-kinase 2 |
| chr17_-_34847013 | 0.48 |
ENSMUST00000166040.9
|
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr2_+_30331839 | 0.48 |
ENSMUST00000131476.8
|
Ptpa
|
protein phosphatase 2 protein activator |
| chr19_-_11816583 | 0.47 |
ENSMUST00000214887.2
|
Olfr1417
|
olfactory receptor 1417 |
| chr11_-_51579441 | 0.46 |
ENSMUST00000007921.9
|
0610009B22Rik
|
RIKEN cDNA 0610009B22 gene |
| chr10_+_73657753 | 0.45 |
ENSMUST00000134009.9
ENSMUST00000177420.7 ENSMUST00000125006.9 |
Pcdh15
|
protocadherin 15 |
| chr14_-_40730180 | 0.45 |
ENSMUST00000136661.8
|
Prxl2a
|
peroxiredoxin like 2A |
| chr5_-_136275407 | 0.45 |
ENSMUST00000196245.2
|
Sh2b2
|
SH2B adaptor protein 2 |
| chr15_-_5137951 | 0.44 |
ENSMUST00000141020.2
|
Card6
|
caspase recruitment domain family, member 6 |
| chr6_+_66706366 | 0.44 |
ENSMUST00000226974.2
ENSMUST00000226311.2 ENSMUST00000228791.2 |
Vmn1r37
|
vomeronasal 1 receptor 37 |
| chr17_-_34847354 | 0.44 |
ENSMUST00000167097.9
|
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr17_+_23819818 | 0.42 |
ENSMUST00000095595.9
ENSMUST00000115509.8 ENSMUST00000120967.8 ENSMUST00000148062.8 ENSMUST00000129227.8 |
Zscan10
|
zinc finger and SCAN domain containing 10 |
| chr7_+_28222261 | 0.41 |
ENSMUST00000078364.4
|
Ifnl3
|
interferon lambda 3 |
| chr11_-_83959999 | 0.40 |
ENSMUST00000138208.2
|
Dusp14
|
dual specificity phosphatase 14 |
| chr2_-_152857239 | 0.39 |
ENSMUST00000028972.9
|
Pdrg1
|
p53 and DNA damage regulated 1 |
| chr7_-_141729738 | 0.39 |
ENSMUST00000190456.2
ENSMUST00000067978.6 |
Krtap5-2
|
keratin associated protein 5-2 |
| chr14_+_54439232 | 0.39 |
ENSMUST00000103724.2
|
Traj17
|
T cell receptor alpha joining 17 |
| chr6_-_67468831 | 0.38 |
ENSMUST00000118364.2
|
Il23r
|
interleukin 23 receptor |
| chr9_-_65427475 | 0.36 |
ENSMUST00000217646.2
|
Ankdd1a
|
ankyrin repeat and death domain containing 1A |
| chr17_-_52117894 | 0.33 |
ENSMUST00000176669.8
|
Satb1
|
special AT-rich sequence binding protein 1 |
| chr3_-_84212069 | 0.33 |
ENSMUST00000107692.8
|
Trim2
|
tripartite motif-containing 2 |
| chr1_+_133254950 | 0.32 |
ENSMUST00000178033.5
|
Kiss1
|
KiSS-1 metastasis-suppressor |
| chr7_-_28209880 | 0.31 |
ENSMUST00000081684.3
|
Ifnl2
|
interferon lambda 2 |
| chr1_+_75119472 | 0.31 |
ENSMUST00000189650.7
|
Retreg2
|
reticulophagy regulator family member 2 |
| chr3_-_89671888 | 0.31 |
ENSMUST00000200558.5
ENSMUST00000029562.5 |
Chrnb2
|
cholinergic receptor, nicotinic, beta polypeptide 2 (neuronal) |
| chr1_+_86631510 | 0.30 |
ENSMUST00000168237.8
ENSMUST00000065694.8 |
Dis3l2
|
DIS3 like 3'-5' exoribonuclease 2 |
| chr1_+_75119809 | 0.29 |
ENSMUST00000186037.7
ENSMUST00000187901.2 |
Retreg2
|
reticulophagy regulator family member 2 |
| chr2_-_48839276 | 0.29 |
ENSMUST00000028098.11
|
Orc4
|
origin recognition complex, subunit 4 |
| chrX_-_101295787 | 0.29 |
ENSMUST00000050551.10
|
Cited1
|
Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1 |
| chr2_-_91540864 | 0.27 |
ENSMUST00000028678.9
ENSMUST00000076803.12 |
Atg13
|
autophagy related 13 |
| chr2_+_36958319 | 0.27 |
ENSMUST00000120704.2
|
Olfr360
|
olfactory receptor 360 |
| chr1_-_20854490 | 0.27 |
ENSMUST00000039046.10
|
Il17f
|
interleukin 17F |
| chr4_+_90111862 | 0.27 |
ENSMUST00000080541.5
|
Zfp352
|
zinc finger protein 352 |
| chr6_+_116531196 | 0.26 |
ENSMUST00000217313.2
ENSMUST00000214699.2 ENSMUST00000215846.2 ENSMUST00000203867.2 |
Olfr214
|
olfactory receptor 214 |
| chr11_+_40624763 | 0.25 |
ENSMUST00000127382.2
|
Nudcd2
|
NudC domain containing 2 |
| chr2_-_151528259 | 0.25 |
ENSMUST00000149319.2
|
Tmem74bos
|
transmembrane 74B, opposite strand |
| chr7_+_141765529 | 0.25 |
ENSMUST00000097943.2
|
Gm7579
|
predicted gene 7579 |
| chr7_+_105017437 | 0.25 |
ENSMUST00000098152.3
ENSMUST00000217827.2 |
Olfr692
|
olfactory receptor 692 |
| chr9_+_32027335 | 0.25 |
ENSMUST00000174641.8
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr1_+_133251701 | 0.23 |
ENSMUST00000195286.5
|
Gm28040
|
predicted gene, 28040 |
| chr17_+_34124078 | 0.23 |
ENSMUST00000172817.2
|
Smim40
|
small integral membrane protein 40 |
| chr14_+_105496147 | 0.23 |
ENSMUST00000138283.2
|
Ndfip2
|
Nedd4 family interacting protein 2 |
| chr2_-_86180622 | 0.22 |
ENSMUST00000099894.5
ENSMUST00000213564.3 |
Olfr1055
|
olfactory receptor 1055 |
| chr11_-_58435764 | 0.21 |
ENSMUST00000108822.8
|
Olfr329
|
olfactory receptor 329 |
| chr7_-_126807581 | 0.21 |
ENSMUST00000120705.3
|
Tbc1d10b
|
TBC1 domain family, member 10b |
| chr1_+_143615294 | 0.20 |
ENSMUST00000185362.7
ENSMUST00000111957.10 ENSMUST00000145571.2 |
Glrx2
|
glutaredoxin 2 (thioltransferase) |
| chr12_+_3941728 | 0.20 |
ENSMUST00000172689.8
ENSMUST00000111186.8 |
Dnmt3a
|
DNA methyltransferase 3A |
| chr8_-_85807308 | 0.20 |
ENSMUST00000093357.12
|
Wdr83
|
WD repeat domain containing 83 |
| chr2_-_111062182 | 0.20 |
ENSMUST00000099620.5
|
Olfr1275
|
olfactory receptor 1275 |
| chr4_+_54945038 | 0.19 |
ENSMUST00000133895.8
|
Zfp462
|
zinc finger protein 462 |
| chr6_-_66537080 | 0.19 |
ENSMUST00000079584.3
ENSMUST00000227014.2 |
Vmn1r32
|
vomeronasal 1 receptor 32 |
| chr11_+_102775991 | 0.18 |
ENSMUST00000100369.4
|
Fam187a
|
family with sequence similarity 187, member A |
| chr7_+_112806672 | 0.18 |
ENSMUST00000047321.9
ENSMUST00000210074.2 ENSMUST00000210238.2 |
Arntl
|
aryl hydrocarbon receptor nuclear translocator-like |
| chr4_-_20778847 | 0.18 |
ENSMUST00000102998.4
|
Nkain3
|
Na+/K+ transporting ATPase interacting 3 |
| chr14_+_54405047 | 0.16 |
ENSMUST00000103693.2
|
Traj50
|
T cell receptor alpha joining 50 |
| chr14_+_53100756 | 0.16 |
ENSMUST00000103616.5
ENSMUST00000186370.2 |
Trav15d-1-dv6d-1
|
T cell receptor alpha variable 15D-1-DV6D-1 |
| chr8_-_25086976 | 0.16 |
ENSMUST00000033956.7
|
Ido1
|
indoleamine 2,3-dioxygenase 1 |
| chr15_+_58287305 | 0.15 |
ENSMUST00000037270.5
|
Fam91a1
|
family with sequence similarity 91, member A1 |
| chr7_+_126895531 | 0.15 |
ENSMUST00000170971.8
|
Itgal
|
integrin alpha L |
| chr2_+_91541245 | 0.15 |
ENSMUST00000142692.2
ENSMUST00000090608.6 |
Harbi1
|
harbinger transposase derived 1 |
| chrX_+_7473338 | 0.14 |
ENSMUST00000115726.9
ENSMUST00000133637.8 ENSMUST00000115725.9 |
Cacna1f
|
calcium channel, voltage-dependent, alpha 1F subunit |
| chr2_-_148613409 | 0.14 |
ENSMUST00000028934.3
|
Cst11
|
cystatin 11 |
| chr2_+_61423469 | 0.14 |
ENSMUST00000112494.2
|
Tank
|
TRAF family member-associated Nf-kappa B activator |
| chr9_+_38260381 | 0.14 |
ENSMUST00000076504.2
|
Olfr898
|
olfactory receptor 898 |
| chr2_-_48839218 | 0.14 |
ENSMUST00000090976.10
ENSMUST00000149679.8 |
Orc4
|
origin recognition complex, subunit 4 |
| chr1_+_23800830 | 0.13 |
ENSMUST00000144602.2
|
B3gat2
|
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) |
| chr9_-_54554483 | 0.13 |
ENSMUST00000128163.8
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr4_-_143693520 | 0.12 |
ENSMUST00000105763.2
|
Pramel28
|
PRAME like 28 |
| chr17_+_38110779 | 0.12 |
ENSMUST00000215168.2
ENSMUST00000216478.2 |
Olfr124
|
olfactory receptor 124 |
| chr8_-_95405234 | 0.12 |
ENSMUST00000213043.2
|
Pllp
|
plasma membrane proteolipid |
| chr5_-_144944838 | 0.12 |
ENSMUST00000116454.10
|
Kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr9_+_54771064 | 0.12 |
ENSMUST00000034843.9
|
Ireb2
|
iron responsive element binding protein 2 |
| chr9_+_19495807 | 0.10 |
ENSMUST00000061693.5
|
Olfr855
|
olfactory receptor 855 |
| chr1_+_171723231 | 0.10 |
ENSMUST00000097466.3
|
Gm10521
|
predicted gene 10521 |
| chr9_-_78263025 | 0.09 |
ENSMUST00000125479.8
|
Gsta2
|
glutathione S-transferase, alpha 2 (Yc2) |
| chr17_+_43671314 | 0.09 |
ENSMUST00000226087.2
|
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr2_-_5867734 | 0.09 |
ENSMUST00000071016.3
|
Gm13199
|
predicted gene 13199 |
| chr2_+_91541197 | 0.09 |
ENSMUST00000128140.2
ENSMUST00000140183.2 |
Harbi1
|
harbinger transposase derived 1 |
| chr2_+_111156865 | 0.08 |
ENSMUST00000208176.3
|
Olfr1281
|
olfactory receptor 1281 |
| chr6_-_124519240 | 0.08 |
ENSMUST00000159463.8
ENSMUST00000162844.2 ENSMUST00000160505.8 ENSMUST00000162443.8 |
C1s1
|
complement component 1, s subcomponent 1 |
| chr3_+_53371086 | 0.08 |
ENSMUST00000058577.5
|
Proser1
|
proline and serine rich 1 |
| chr5_-_145020840 | 0.07 |
ENSMUST00000151196.2
|
Kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr7_+_106513407 | 0.06 |
ENSMUST00000098140.2
|
Olfr1532-ps1
|
olfactory receptor 1532, pseudogene 1 |
| chr14_-_60434622 | 0.06 |
ENSMUST00000131670.3
|
Atp8a2
|
ATPase, aminophospholipid transporter-like, class I, type 8A, member 2 |
| chr6_-_93890520 | 0.05 |
ENSMUST00000203688.3
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr2_-_111965322 | 0.05 |
ENSMUST00000213696.2
|
Olfr1316
|
olfactory receptor 1316 |
| chr13_-_21722197 | 0.05 |
ENSMUST00000168629.2
ENSMUST00000218154.2 |
Olfr1366
|
olfactory receptor 1366 |
| chr9_+_77661808 | 0.05 |
ENSMUST00000034905.9
|
Gclc
|
glutamate-cysteine ligase, catalytic subunit |
| chr15_-_11996084 | 0.05 |
ENSMUST00000022816.15
|
Sub1
|
SUB1 homolog, transcriptional regulator |
| chr8_-_3674993 | 0.05 |
ENSMUST00000142431.8
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chrX_+_41239872 | 0.05 |
ENSMUST00000123245.8
|
Stag2
|
stromal antigen 2 |
| chr7_-_126522014 | 0.04 |
ENSMUST00000134134.3
ENSMUST00000119781.8 ENSMUST00000121612.4 |
Tmem219
|
transmembrane protein 219 |
| chr13_+_21679387 | 0.03 |
ENSMUST00000104942.2
|
AK157302
|
cDNA sequence AK157302 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 7.6 | GO:0015886 | heme transport(GO:0015886) positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 1.1 | 9.5 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 1.0 | 4.1 | GO:0030974 | thiamine pyrophosphate transport(GO:0030974) |
| 0.8 | 2.4 | GO:0038190 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 0.6 | 7.0 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.6 | 2.4 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.5 | 1.4 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.5 | 2.3 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.4 | 1.8 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.4 | 1.6 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.4 | 0.8 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.4 | 1.5 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.3 | 1.4 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.3 | 2.0 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.3 | 2.3 | GO:0009750 | response to fructose(GO:0009750) |
| 0.2 | 1.7 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.2 | 1.8 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.2 | 0.8 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.2 | 0.7 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.2 | 2.7 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.2 | 0.5 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.1 | 0.6 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 3.4 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 1.6 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 1.3 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 0.7 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 | 1.3 | GO:0098970 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.1 | 1.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 0.7 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 | 1.1 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 2.7 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.3 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) generation of ovulation cycle rhythm(GO:0060112) |
| 0.1 | 0.3 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.1 | 1.4 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.1 | 1.9 | GO:0009251 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 1.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.8 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.8 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.3 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.1 | 0.2 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 1.6 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.3 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 1.2 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.7 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 0.4 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 1.3 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.1 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.2 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 2.4 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 1.4 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 1.3 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 0.3 | GO:0045414 | regulation of interleukin-8 biosynthetic process(GO:0045414) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.9 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 2.2 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 0.4 | GO:0002827 | positive regulation of T-helper 1 type immune response(GO:0002827) |
| 0.0 | 0.9 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.3 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 1.6 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 3.6 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.0 | 1.2 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.2 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 1.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0003011 | involuntary skeletal muscle contraction(GO:0003011) |
| 0.0 | 0.3 | GO:0002385 | organ or tissue specific immune response(GO:0002251) mucosal immune response(GO:0002385) |
| 0.0 | 0.5 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.3 | GO:0098780 | response to mitochondrial depolarisation(GO:0098780) |
| 0.0 | 2.7 | GO:0007623 | circadian rhythm(GO:0007623) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.2 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.5 | 1.4 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.2 | 2.7 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 1.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 4.1 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 0.7 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.1 | 1.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 6.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.4 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 3.8 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 3.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 7.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.0 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 4.3 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.7 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.8 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.0 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.3 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 5.3 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.2 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 1.3 | 7.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.8 | 2.4 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.7 | 2.2 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.6 | 4.1 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 0.5 | 1.9 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.5 | 1.4 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.3 | 2.7 | GO:0032052 | long-chain fatty acid transporter activity(GO:0005324) bile acid binding(GO:0032052) |
| 0.3 | 2.0 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.2 | 2.3 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.2 | 1.4 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 1.3 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 0.6 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.1 | 1.8 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.1 | 2.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.5 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.1 | 6.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 4.0 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.1 | 2.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.9 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 6.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 2.3 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.1 | 2.7 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.4 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 0.4 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 1.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.2 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.1 | 3.4 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 1.8 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 7.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 2.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 1.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.7 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.7 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 5.8 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.3 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 1.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 1.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.8 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 8.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 11.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 7.6 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 1.5 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.8 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 2.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 2.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 2.3 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.1 | 3.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.4 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 1.6 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.4 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.7 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 1.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.4 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.5 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |