Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Runx2_Bcl11a
Z-value: 2.88
Transcription factors associated with Runx2_Bcl11a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
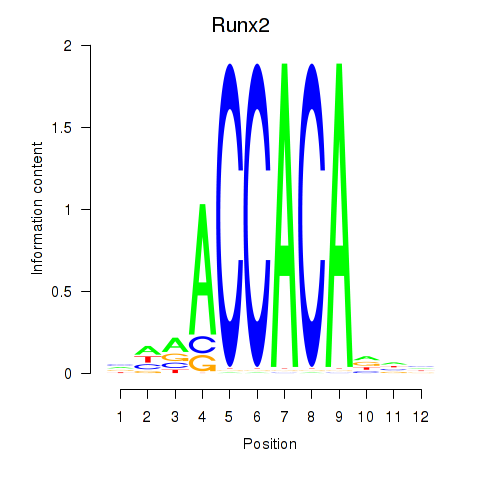
Runx2
|
ENSMUSG00000039153.18 | Runx2 |
|
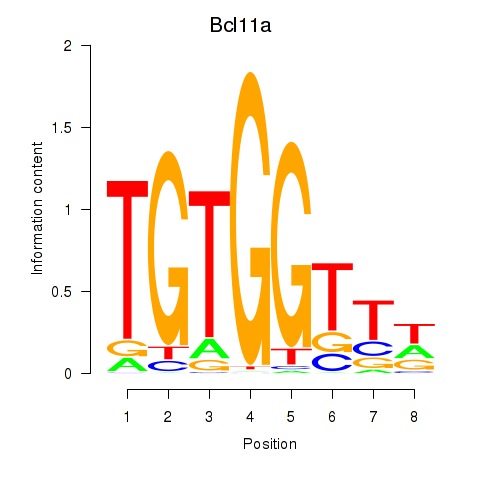
Bcl11a
|
ENSMUSG00000000861.16 | Bcl11a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Runx2 | mm39_v1_chr17_-_45125537_45125684 | 0.41 | 3.5e-04 | Click! |
| Bcl11a | mm39_v1_chr11_+_24028173_24028220 | 0.18 | 1.2e-01 | Click! |
Activity profile of Runx2_Bcl11a motif
Sorted Z-values of Runx2_Bcl11a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Runx2_Bcl11a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_35782412 | 33.11 |
ENSMUST00000025211.6
|
Mzb1
|
marginal zone B and B1 cell-specific protein 1 |
| chr11_-_106205320 | 30.15 |
ENSMUST00000167143.2
|
Cd79b
|
CD79B antigen |
| chr7_-_103477126 | 28.41 |
ENSMUST00000023934.8
|
Hbb-bs
|
hemoglobin, beta adult s chain |
| chr3_+_103767581 | 28.30 |
ENSMUST00000029433.9
|
Ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr7_+_142559414 | 26.28 |
ENSMUST00000082008.12
ENSMUST00000105925.8 ENSMUST00000105924.8 |
Tspan32
|
tetraspanin 32 |
| chrX_-_100311824 | 26.16 |
ENSMUST00000033664.14
|
Il2rg
|
interleukin 2 receptor, gamma chain |
| chr3_-_59009231 | 25.71 |
ENSMUST00000085040.5
|
Gpr171
|
G protein-coupled receptor 171 |
| chr6_-_87510200 | 25.61 |
ENSMUST00000113637.9
ENSMUST00000071024.7 |
Arhgap25
|
Rho GTPase activating protein 25 |
| chr7_+_142559375 | 24.59 |
ENSMUST00000075172.12
ENSMUST00000105923.8 |
Tspan32
|
tetraspanin 32 |
| chr7_+_142558783 | 23.82 |
ENSMUST00000009396.13
|
Tspan32
|
tetraspanin 32 |
| chr6_+_41248311 | 22.74 |
ENSMUST00000103281.3
|
Trbv29
|
T cell receptor beta, variable 29 |
| chr7_+_142558837 | 22.70 |
ENSMUST00000207211.2
|
Tspan32
|
tetraspanin 32 |
| chr11_-_79414542 | 22.62 |
ENSMUST00000179322.2
|
Evi2b
|
ecotropic viral integration site 2b |
| chrX_-_7956682 | 20.87 |
ENSMUST00000033505.7
|
Was
|
Wiskott-Aldrich syndrome |
| chr15_-_78377926 | 20.34 |
ENSMUST00000163494.3
|
Il2rb
|
interleukin 2 receptor, beta chain |
| chr6_+_41118120 | 20.31 |
ENSMUST00000103273.3
|
Trbv15
|
T cell receptor beta, variable 15 |
| chr5_-_113957318 | 19.87 |
ENSMUST00000201194.4
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr8_-_3744167 | 19.28 |
ENSMUST00000005678.6
|
Fcer2a
|
Fc receptor, IgE, low affinity II, alpha polypeptide |
| chr7_-_140596811 | 19.09 |
ENSMUST00000081924.5
|
Ifitm6
|
interferon induced transmembrane protein 6 |
| chr2_-_58050494 | 18.78 |
ENSMUST00000028175.7
|
Cytip
|
cytohesin 1 interacting protein |
| chr14_-_56499690 | 18.65 |
ENSMUST00000015581.6
|
Gzmb
|
granzyme B |
| chr8_-_110305672 | 16.72 |
ENSMUST00000074898.8
|
Hp
|
haptoglobin |
| chr16_-_18884431 | 16.48 |
ENSMUST00000200235.2
|
Iglc3
|
immunoglobulin lambda constant 3 |
| chr1_+_135060994 | 16.47 |
ENSMUST00000167080.3
|
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr5_+_90916370 | 16.01 |
ENSMUST00000031319.8
|
Ppbp
|
pro-platelet basic protein |
| chr19_+_3985186 | 16.00 |
ENSMUST00000165711.3
ENSMUST00000162708.7 ENSMUST00000236920.2 |
Unc93b1
|
unc-93 homolog B1, TLR signaling regulator |
| chr11_-_46280336 | 15.95 |
ENSMUST00000020664.13
|
Itk
|
IL2 inducible T cell kinase |
| chr15_-_9529898 | 15.30 |
ENSMUST00000228782.2
ENSMUST00000003981.6 |
Il7r
|
interleukin 7 receptor |
| chr16_+_23338960 | 15.19 |
ENSMUST00000211460.2
ENSMUST00000210658.2 ENSMUST00000209198.2 ENSMUST00000210371.2 ENSMUST00000211499.2 ENSMUST00000210795.2 ENSMUST00000209422.2 |
Gm45338
Rtp4
|
predicted gene 45338 receptor transporter protein 4 |
| chr5_-_113957362 | 15.07 |
ENSMUST00000202555.2
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr12_-_112824506 | 15.01 |
ENSMUST00000021729.9
|
Gpr132
|
G protein-coupled receptor 132 |
| chr1_+_135060431 | 14.98 |
ENSMUST00000187985.7
ENSMUST00000049449.11 |
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr5_+_86219593 | 14.91 |
ENSMUST00000198435.5
ENSMUST00000031171.9 |
Stap1
|
signal transducing adaptor family member 1 |
| chr16_-_92622972 | 14.66 |
ENSMUST00000023673.14
|
Runx1
|
runt related transcription factor 1 |
| chr11_-_106278892 | 14.46 |
ENSMUST00000106813.9
ENSMUST00000141146.2 |
Icam2
|
intercellular adhesion molecule 2 |
| chrX_-_100312629 | 14.37 |
ENSMUST00000117736.2
|
Gm20489
|
predicted gene 20489 |
| chr11_-_46280298 | 14.30 |
ENSMUST00000109237.9
|
Itk
|
IL2 inducible T cell kinase |
| chr11_-_87766350 | 14.22 |
ENSMUST00000049768.4
|
Epx
|
eosinophil peroxidase |
| chr4_+_134847949 | 14.14 |
ENSMUST00000056977.14
|
Runx3
|
runt related transcription factor 3 |
| chr7_+_142559475 | 14.07 |
ENSMUST00000143512.3
|
Tspan32
|
tetraspanin 32 |
| chr16_-_45940602 | 13.63 |
ENSMUST00000023336.10
|
Cd96
|
CD96 antigen |
| chr13_-_20008397 | 12.69 |
ENSMUST00000222664.2
ENSMUST00000065335.3 |
Gpr141
|
G protein-coupled receptor 141 |
| chr11_-_46280281 | 12.68 |
ENSMUST00000101306.4
|
Itk
|
IL2 inducible T cell kinase |
| chr1_+_170104889 | 12.65 |
ENSMUST00000179976.3
|
Sh2d1b1
|
SH2 domain containing 1B1 |
| chr7_+_44221791 | 12.55 |
ENSMUST00000002274.10
|
Napsa
|
napsin A aspartic peptidase |
| chr6_-_69521891 | 12.46 |
ENSMUST00000103356.4
|
Igkv4-57-1
|
immunoglobulin kappa variable 4-57-1 |
| chr13_-_113237505 | 12.33 |
ENSMUST00000224282.2
ENSMUST00000023897.7 |
Gzma
|
granzyme A |
| chr6_+_41128636 | 11.74 |
ENSMUST00000103274.4
|
Trbv16
|
T cell receptor beta, variable 16 |
| chr7_+_43086432 | 11.58 |
ENSMUST00000070518.4
|
Nkg7
|
natural killer cell group 7 sequence |
| chr15_+_80456775 | 11.38 |
ENSMUST00000229980.2
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr11_-_69786324 | 11.35 |
ENSMUST00000001631.7
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chrX_+_106299484 | 11.35 |
ENSMUST00000101294.9
ENSMUST00000118820.8 ENSMUST00000120971.8 |
Gpr174
|
G protein-coupled receptor 174 |
| chr6_+_87755046 | 11.32 |
ENSMUST00000032133.5
|
Gp9
|
glycoprotein 9 (platelet) |
| chr19_-_20368029 | 11.21 |
ENSMUST00000235280.2
|
Anxa1
|
annexin A1 |
| chrX_-_56384089 | 10.96 |
ENSMUST00000033468.11
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr2_-_32277773 | 10.85 |
ENSMUST00000050785.14
|
Lcn2
|
lipocalin 2 |
| chr11_-_83540175 | 10.82 |
ENSMUST00000001008.6
|
Ccl3
|
chemokine (C-C motif) ligand 3 |
| chr6_+_123100382 | 10.71 |
ENSMUST00000032248.8
|
Clec4a2
|
C-type lectin domain family 4, member a2 |
| chr7_-_140845156 | 10.60 |
ENSMUST00000123525.2
|
Irf7
|
interferon regulatory factor 7 |
| chrX_+_158480304 | 10.50 |
ENSMUST00000123433.8
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chrX_+_106192510 | 10.47 |
ENSMUST00000147521.8
ENSMUST00000167673.2 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr15_-_66673425 | 10.45 |
ENSMUST00000168589.8
|
Sla
|
src-like adaptor |
| chr7_+_24069680 | 10.40 |
ENSMUST00000205428.2
ENSMUST00000171904.3 ENSMUST00000205626.2 |
Kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr6_+_123100272 | 10.27 |
ENSMUST00000041779.13
|
Clec4a2
|
C-type lectin domain family 4, member a2 |
| chr1_-_162687488 | 10.27 |
ENSMUST00000134098.8
ENSMUST00000111518.3 |
Fmo1
|
flavin containing monooxygenase 1 |
| chr7_+_43086554 | 10.20 |
ENSMUST00000206741.2
|
Nkg7
|
natural killer cell group 7 sequence |
| chr5_+_105667254 | 9.92 |
ENSMUST00000067924.13
ENSMUST00000150981.2 |
Lrrc8c
|
leucine rich repeat containing 8 family, member C |
| chr5_-_105287405 | 9.91 |
ENSMUST00000100961.5
ENSMUST00000031235.13 ENSMUST00000197799.2 ENSMUST00000199629.2 ENSMUST00000196677.5 ENSMUST00000100962.8 |
Gbp9
Gbp8
Gbp4
|
guanylate-binding protein 9 guanylate-binding protein 8 guanylate binding protein 4 |
| chr8_+_89020845 | 9.91 |
ENSMUST00000098521.4
|
Adcy7
|
adenylate cyclase 7 |
| chr6_-_48747767 | 9.84 |
ENSMUST00000204036.2
|
Gimap3
|
GTPase, IMAP family member 3 |
| chr17_+_47906985 | 9.75 |
ENSMUST00000182539.8
|
Ccnd3
|
cyclin D3 |
| chr18_-_62313019 | 9.67 |
ENSMUST00000053640.5
|
Adrb2
|
adrenergic receptor, beta 2 |
| chr10_-_62343516 | 9.62 |
ENSMUST00000020271.13
|
Srgn
|
serglycin |
| chr11_+_48967411 | 9.56 |
ENSMUST00000109202.3
|
Ifi47
|
interferon gamma inducible protein 47 |
| chr17_+_33857030 | 9.41 |
ENSMUST00000052079.8
|
Pram1
|
PML-RAR alpha-regulated adaptor molecule 1 |
| chr15_+_6599001 | 9.39 |
ENSMUST00000227175.2
|
Fyb
|
FYN binding protein |
| chr9_+_72345267 | 9.36 |
ENSMUST00000183809.8
|
Mns1
|
meiosis-specific nuclear structural protein 1 |
| chrX_-_100777806 | 9.29 |
ENSMUST00000056614.7
|
Cxcr3
|
chemokine (C-X-C motif) receptor 3 |
| chr15_-_97742134 | 9.21 |
ENSMUST00000119670.8
ENSMUST00000116409.9 |
Hdac7
|
histone deacetylase 7 |
| chr5_-_107873883 | 9.17 |
ENSMUST00000159263.3
|
Gfi1
|
growth factor independent 1 transcription repressor |
| chr17_+_35235552 | 9.14 |
ENSMUST00000007245.8
ENSMUST00000172499.2 |
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr3_-_135313982 | 9.14 |
ENSMUST00000132668.8
|
Nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 1, p105 |
| chr16_-_16681839 | 9.07 |
ENSMUST00000100136.4
|
Igll1
|
immunoglobulin lambda-like polypeptide 1 |
| chr5_-_113970664 | 9.06 |
ENSMUST00000199109.2
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr19_-_10807409 | 9.03 |
ENSMUST00000080292.12
|
Cd6
|
CD6 antigen |
| chr1_-_172418058 | 9.03 |
ENSMUST00000065679.8
|
Slamf8
|
SLAM family member 8 |
| chr4_+_135899678 | 8.98 |
ENSMUST00000061721.6
|
E2f2
|
E2F transcription factor 2 |
| chr1_-_192880260 | 8.93 |
ENSMUST00000161367.2
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr11_+_95733109 | 8.91 |
ENSMUST00000107714.9
ENSMUST00000107711.8 |
Gngt2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr11_-_52173391 | 8.83 |
ENSMUST00000086844.10
|
Tcf7
|
transcription factor 7, T cell specific |
| chr7_-_44888220 | 8.80 |
ENSMUST00000210372.2
ENSMUST00000209779.2 ENSMUST00000098461.10 ENSMUST00000211373.2 |
Cd37
|
CD37 antigen |
| chr19_-_10807285 | 8.76 |
ENSMUST00000039043.15
|
Cd6
|
CD6 antigen |
| chr17_+_56056977 | 8.71 |
ENSMUST00000025004.7
|
Adgre4
|
adhesion G protein-coupled receptor E4 |
| chr19_+_4175934 | 8.56 |
ENSMUST00000169055.10
ENSMUST00000135257.11 ENSMUST00000138593.10 ENSMUST00000140267.10 ENSMUST00000137579.6 ENSMUST00000139987.10 ENSMUST00000150627.11 ENSMUST00000151401.11 ENSMUST00000153654.11 |
Tmem134
|
transmembrane protein 134 |
| chr17_+_8463886 | 8.55 |
ENSMUST00000231545.2
|
Ccr6
|
chemokine (C-C motif) receptor 6 |
| chr1_-_164763091 | 8.51 |
ENSMUST00000027860.8
|
Xcl1
|
chemokine (C motif) ligand 1 |
| chr10_+_28544356 | 8.35 |
ENSMUST00000060409.13
ENSMUST00000056097.11 ENSMUST00000105516.9 |
Themis
|
thymocyte selection associated |
| chr7_-_100512579 | 8.33 |
ENSMUST00000142885.2
ENSMUST00000008462.11 |
Relt
|
RELT tumor necrosis factor receptor |
| chr15_-_97742045 | 8.31 |
ENSMUST00000120683.8
|
Hdac7
|
histone deacetylase 7 |
| chr16_-_19060440 | 8.16 |
ENSMUST00000103751.3
|
Iglv3
|
immunoglobulin lambda variable 3 |
| chr1_+_107456731 | 8.16 |
ENSMUST00000182198.8
|
Serpinb10
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10 |
| chrX_+_149330371 | 7.86 |
ENSMUST00000066337.13
ENSMUST00000112715.2 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr9_-_70842090 | 7.86 |
ENSMUST00000034731.10
|
Lipc
|
lipase, hepatic |
| chr16_-_18880821 | 7.85 |
ENSMUST00000200568.2
|
Iglc1
|
immunoglobulin lambda constant 1 |
| chr13_-_55169100 | 7.74 |
ENSMUST00000148221.8
ENSMUST00000052949.13 |
Hk3
|
hexokinase 3 |
| chr9_+_108880221 | 7.71 |
ENSMUST00000200629.5
ENSMUST00000200515.5 ENSMUST00000197689.5 ENSMUST00000196954.5 ENSMUST00000198376.5 ENSMUST00000197483.5 ENSMUST00000198295.5 |
Shisa5
|
shisa family member 5 |
| chr17_+_34457868 | 7.70 |
ENSMUST00000095342.11
ENSMUST00000167280.8 ENSMUST00000236838.2 |
H2-Ob
|
histocompatibility 2, O region beta locus |
| chr1_-_173569301 | 7.59 |
ENSMUST00000042610.15
ENSMUST00000127730.2 |
Ifi207
|
interferon activated gene 207 |
| chr1_-_173859321 | 7.55 |
ENSMUST00000059226.7
|
Ifi205
|
interferon activated gene 205 |
| chr6_+_123099615 | 7.49 |
ENSMUST00000161636.8
ENSMUST00000161365.8 |
Clec4a2
|
C-type lectin domain family 4, member a2 |
| chr12_-_4924341 | 7.46 |
ENSMUST00000137337.8
ENSMUST00000045921.14 |
Mfsd2b
|
major facilitator superfamily domain containing 2B |
| chr3_-_15397325 | 7.45 |
ENSMUST00000108361.2
|
Gm9733
|
predicted gene 9733 |
| chr1_-_192883743 | 7.40 |
ENSMUST00000043550.11
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr13_-_55169000 | 7.32 |
ENSMUST00000153665.8
|
Hk3
|
hexokinase 3 |
| chrX_-_9335525 | 7.32 |
ENSMUST00000015484.10
|
Cybb
|
cytochrome b-245, beta polypeptide |
| chr2_-_73316053 | 7.23 |
ENSMUST00000102680.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr4_-_42773987 | 7.16 |
ENSMUST00000095114.5
|
Ccl21a
|
chemokine (C-C motif) ligand 21A (serine) |
| chr17_-_45125537 | 7.13 |
ENSMUST00000113571.10
|
Runx2
|
runt related transcription factor 2 |
| chr8_+_84852609 | 7.08 |
ENSMUST00000093380.5
|
Podnl1
|
podocan-like 1 |
| chr7_-_140597465 | 6.93 |
ENSMUST00000211330.2
|
Ifitm6
|
interferon induced transmembrane protein 6 |
| chr17_+_34544632 | 6.88 |
ENSMUST00000050325.10
|
H2-Eb2
|
histocompatibility 2, class II antigen E beta2 |
| chr17_+_37180437 | 6.87 |
ENSMUST00000060524.11
|
Trim10
|
tripartite motif-containing 10 |
| chr5_+_145063568 | 6.86 |
ENSMUST00000138922.2
|
Arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr17_-_34506744 | 6.85 |
ENSMUST00000174751.2
ENSMUST00000040655.14 |
H2-Aa
|
histocompatibility 2, class II antigen A, alpha |
| chr19_-_10807220 | 6.85 |
ENSMUST00000174176.3
|
Cd6
|
CD6 antigen |
| chr2_-_101479846 | 6.79 |
ENSMUST00000078494.6
ENSMUST00000160722.8 ENSMUST00000160037.8 |
Rag1
Iftap
|
recombination activating 1 intraflagellar transport associated protein |
| chr6_+_70700207 | 6.77 |
ENSMUST00000103407.3
ENSMUST00000199487.2 |
Igkj3
|
immunoglobulin kappa joining 3 |
| chr7_-_43182595 | 6.75 |
ENSMUST00000205503.2
|
Cd33
|
CD33 antigen |
| chr6_-_69626340 | 6.69 |
ENSMUST00000198328.2
|
Igkv4-53
|
immunoglobulin kappa variable 4-53 |
| chr14_-_122153185 | 6.69 |
ENSMUST00000055475.9
|
Gpr18
|
G protein-coupled receptor 18 |
| chr17_+_31783708 | 6.66 |
ENSMUST00000097352.11
ENSMUST00000237248.2 ENSMUST00000235869.2 ENSMUST00000175806.9 |
Pknox1
|
Pbx/knotted 1 homeobox |
| chr10_+_79852487 | 6.65 |
ENSMUST00000099501.10
|
Arhgap45
|
Rho GTPase activating protein 45 |
| chr2_-_173060647 | 6.62 |
ENSMUST00000109116.3
ENSMUST00000029018.14 |
Zbp1
|
Z-DNA binding protein 1 |
| chr11_-_16958647 | 6.62 |
ENSMUST00000102881.10
|
Plek
|
pleckstrin |
| chr9_+_51124983 | 6.61 |
ENSMUST00000034554.9
|
Pou2af1
|
POU domain, class 2, associating factor 1 |
| chr14_+_53756760 | 6.60 |
ENSMUST00000103585.4
|
Trav11
|
T cell receptor alpha variable 11 |
| chr10_+_79852750 | 6.60 |
ENSMUST00000105373.8
|
Arhgap45
|
Rho GTPase activating protein 45 |
| chr17_+_35413415 | 6.58 |
ENSMUST00000025262.6
ENSMUST00000173600.2 |
Ltb
|
lymphotoxin B |
| chr9_+_110867807 | 6.56 |
ENSMUST00000197575.2
|
Ltf
|
lactotransferrin |
| chrX_-_12026594 | 6.53 |
ENSMUST00000043441.13
|
Bcor
|
BCL6 interacting corepressor |
| chr9_+_108880158 | 6.50 |
ENSMUST00000198708.5
|
Shisa5
|
shisa family member 5 |
| chr2_+_26969384 | 6.49 |
ENSMUST00000091233.7
|
Adamtsl2
|
ADAMTS-like 2 |
| chr2_-_26561294 | 6.48 |
ENSMUST00000028283.2
|
Lcn4
|
lipocalin 4 |
| chr11_-_120930193 | 6.45 |
ENSMUST00000026159.6
|
Cd7
|
CD7 antigen |
| chr7_-_3848050 | 6.39 |
ENSMUST00000108615.10
ENSMUST00000119469.2 |
Pira2
|
paired-Ig-like receptor A2 |
| chr14_+_53743184 | 6.21 |
ENSMUST00000103583.5
|
Trav10
|
T cell receptor alpha variable 10 |
| chr10_+_118276949 | 6.16 |
ENSMUST00000068592.5
|
Ifng
|
interferon gamma |
| chr6_+_41098273 | 6.15 |
ENSMUST00000103270.4
|
Trbv13-2
|
T cell receptor beta, variable 13-2 |
| chr1_-_173740467 | 6.14 |
ENSMUST00000009340.10
|
Ifi211
|
interferon activated gene 211 |
| chr11_+_82955917 | 6.14 |
ENSMUST00000038038.8
|
Slfn2
|
schlafen 2 |
| chr6_-_130363837 | 6.13 |
ENSMUST00000032288.6
|
Klra1
|
killer cell lectin-like receptor, subfamily A, member 1 |
| chr10_+_61007733 | 6.10 |
ENSMUST00000122261.8
ENSMUST00000121297.8 ENSMUST00000035894.12 |
Tbata
|
thymus, brain and testes associated |
| chr5_-_113968483 | 6.04 |
ENSMUST00000100874.6
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr3_-_86049988 | 6.02 |
ENSMUST00000029722.7
|
Rps3a1
|
ribosomal protein S3A1 |
| chr17_-_35285146 | 5.99 |
ENSMUST00000174190.2
ENSMUST00000097337.8 |
Mpig6b
|
megakaryocyte and platelet inhibitory receptor G6b |
| chr3_+_90421742 | 5.93 |
ENSMUST00000048138.8
|
S100a13
|
S100 calcium binding protein A13 |
| chr7_+_126895463 | 5.92 |
ENSMUST00000106306.9
ENSMUST00000120857.8 |
Itgal
|
integrin alpha L |
| chr1_-_171434882 | 5.89 |
ENSMUST00000111277.2
ENSMUST00000004827.14 |
Ly9
|
lymphocyte antigen 9 |
| chr7_+_28136861 | 5.87 |
ENSMUST00000108292.9
ENSMUST00000108289.8 |
Gmfg
|
glia maturation factor, gamma |
| chr3_+_130906434 | 5.86 |
ENSMUST00000098611.4
|
Lef1
|
lymphoid enhancer binding factor 1 |
| chr11_+_87684299 | 5.86 |
ENSMUST00000020779.11
|
Mpo
|
myeloperoxidase |
| chr7_-_3723381 | 5.76 |
ENSMUST00000078451.7
|
Pirb
|
paired Ig-like receptor B |
| chr7_+_4240697 | 5.74 |
ENSMUST00000117550.2
|
Lilra5
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 |
| chr4_+_114914880 | 5.73 |
ENSMUST00000161601.8
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr8_-_112417633 | 5.71 |
ENSMUST00000034435.7
|
Ctrb1
|
chymotrypsinogen B1 |
| chr16_-_75706161 | 5.71 |
ENSMUST00000114239.9
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr11_+_83637766 | 5.69 |
ENSMUST00000070832.3
|
Wfdc21
|
WAP four-disulfide core domain 21 |
| chr10_+_51356728 | 5.69 |
ENSMUST00000102894.6
ENSMUST00000219661.2 ENSMUST00000219696.2 ENSMUST00000217706.2 |
Lilr4b
Gm49339
|
leukocyte immunoglobulin-like receptor, subfamily B, member 4B predicted gene, 49339 |
| chr19_-_4175837 | 5.64 |
ENSMUST00000121402.2
ENSMUST00000117831.8 |
Aip
|
aryl-hydrocarbon receptor-interacting protein |
| chrX_+_158410528 | 5.62 |
ENSMUST00000073094.10
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr17_+_48623157 | 5.58 |
ENSMUST00000049614.13
|
B430306N03Rik
|
RIKEN cDNA B430306N03 gene |
| chr1_-_38704028 | 5.58 |
ENSMUST00000039827.14
|
Aff3
|
AF4/FMR2 family, member 3 |
| chr7_+_28140352 | 5.48 |
ENSMUST00000078845.13
|
Gmfg
|
glia maturation factor, gamma |
| chr7_-_44888532 | 5.45 |
ENSMUST00000033063.15
|
Cd37
|
CD37 antigen |
| chr1_-_144651157 | 5.44 |
ENSMUST00000027603.4
|
Rgs18
|
regulator of G-protein signaling 18 |
| chr1_-_171434944 | 5.43 |
ENSMUST00000068878.14
|
Ly9
|
lymphocyte antigen 9 |
| chrX_+_41591476 | 5.42 |
ENSMUST00000115070.8
ENSMUST00000153948.2 |
Sh2d1a
|
SH2 domain containing 1A |
| chr1_+_74414354 | 5.42 |
ENSMUST00000187516.7
ENSMUST00000027368.6 |
Slc11a1
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 |
| chr14_+_54198389 | 5.41 |
ENSMUST00000103678.4
|
Trdv2-2
|
T cell receptor delta variable 2-2 |
| chr15_+_6552270 | 5.39 |
ENSMUST00000226412.2
|
Fyb
|
FYN binding protein |
| chr7_+_30264835 | 5.36 |
ENSMUST00000043850.14
|
Igflr1
|
IGF-like family receptor 1 |
| chr4_-_46404224 | 5.36 |
ENSMUST00000107764.9
|
Hemgn
|
hemogen |
| chr7_+_26534730 | 5.35 |
ENSMUST00000005685.15
|
Cyp2a5
|
cytochrome P450, family 2, subfamily a, polypeptide 5 |
| chr10_-_79624758 | 5.28 |
ENSMUST00000020573.13
|
Prss57
|
protease, serine 57 |
| chr14_+_54433095 | 5.26 |
ENSMUST00000103717.3
|
Traj24
|
T cell receptor alpha joining 24 |
| chrX_+_106193060 | 5.24 |
ENSMUST00000125676.8
ENSMUST00000180182.2 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr7_-_3828640 | 5.17 |
ENSMUST00000189095.7
ENSMUST00000094911.5 ENSMUST00000153846.8 ENSMUST00000108619.8 ENSMUST00000108620.8 |
Gm15448
|
predicted gene 15448 |
| chr4_+_114914607 | 5.17 |
ENSMUST00000136946.8
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr13_-_37234213 | 5.16 |
ENSMUST00000164727.8
|
F13a1
|
coagulation factor XIII, A1 subunit |
| chr5_+_90920294 | 5.16 |
ENSMUST00000031320.8
|
Pf4
|
platelet factor 4 |
| chr7_+_126184108 | 5.15 |
ENSMUST00000039522.8
|
Apobr
|
apolipoprotein B receptor |
| chr17_-_45903494 | 5.12 |
ENSMUST00000163492.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr15_+_97259060 | 5.11 |
ENSMUST00000228521.2
ENSMUST00000226495.2 |
Pced1b
|
PC-esterase domain containing 1B |
| chr3_+_145855929 | 5.10 |
ENSMUST00000098524.5
|
Mcoln2
|
mucolipin 2 |
| chr17_+_34482183 | 5.10 |
ENSMUST00000040828.7
ENSMUST00000237342.2 ENSMUST00000237866.2 |
H2-Ab1
|
histocompatibility 2, class II antigen A, beta 1 |
| chrX_+_41591410 | 5.07 |
ENSMUST00000005839.11
|
Sh2d1a
|
SH2 domain containing 1A |
| chr9_+_123603605 | 5.06 |
ENSMUST00000180093.2
|
Ccr9
|
chemokine (C-C motif) receptor 9 |
| chr1_+_172328768 | 5.03 |
ENSMUST00000111228.2
|
Tagln2
|
transgelin 2 |
| chr1_+_36806015 | 5.02 |
ENSMUST00000185871.2
|
Zap70
|
zeta-chain (TCR) associated protein kinase |
| chr7_-_44888465 | 4.98 |
ENSMUST00000210078.2
|
Cd37
|
CD37 antigen |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.3 | 36.8 | GO:2000566 | positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 11.9 | 130.7 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 10.0 | 50.0 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 7.0 | 20.9 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 6.9 | 34.4 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 6.5 | 19.5 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 5.9 | 35.7 | GO:0032831 | positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 5.2 | 15.7 | GO:0002215 | defense response to nematode(GO:0002215) |
| 5.0 | 24.8 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 4.8 | 14.3 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 3.9 | 19.7 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 3.9 | 11.7 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 3.8 | 15.3 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 3.8 | 11.4 | GO:0071846 | actin filament debranching(GO:0071846) |
| 3.7 | 18.6 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 3.7 | 18.3 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) |
| 3.6 | 10.9 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 3.2 | 12.7 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 2.9 | 8.6 | GO:1904156 | DN2 thymocyte differentiation(GO:1904155) DN3 thymocyte differentiation(GO:1904156) |
| 2.8 | 11.3 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 2.8 | 19.3 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 2.6 | 7.9 | GO:0030845 | regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010924) positive regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010925) phospholipase C-inhibiting G-protein coupled receptor signaling pathway(GO:0030845) regulation of cell diameter(GO:0060305) |
| 2.6 | 7.7 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) negative regulation of antigen processing and presentation of peptide antigen(GO:0002584) |
| 2.5 | 7.6 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 2.4 | 9.7 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 2.4 | 9.6 | GO:0033368 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 2.4 | 7.2 | GO:2000547 | regulation of dendritic cell dendrite assembly(GO:2000547) |
| 2.3 | 16.4 | GO:0002370 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 2.3 | 46.6 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 2.2 | 11.2 | GO:0097350 | neutrophil clearance(GO:0097350) |
| 2.2 | 10.9 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 2.1 | 6.3 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 2.0 | 9.8 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 1.9 | 5.8 | GO:0035702 | monocyte homeostasis(GO:0035702) |
| 1.9 | 13.6 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 1.9 | 13.1 | GO:0015862 | uridine transport(GO:0015862) |
| 1.9 | 5.6 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 1.8 | 27.6 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 1.8 | 5.4 | GO:0070839 | divalent metal ion export(GO:0070839) |
| 1.8 | 8.9 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 1.8 | 10.6 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 1.7 | 5.2 | GO:0021664 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 1.7 | 5.1 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 1.7 | 6.8 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 1.7 | 11.7 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 1.7 | 6.6 | GO:0030091 | protein repair(GO:0030091) |
| 1.6 | 8.2 | GO:0090366 | DNA cytosine deamination(GO:0070383) positive regulation of mRNA modification(GO:0090366) |
| 1.6 | 6.6 | GO:0051673 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 1.6 | 6.5 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 1.5 | 1.5 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 1.5 | 9.0 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 1.5 | 5.8 | GO:0001803 | type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 1.4 | 5.6 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 1.4 | 6.8 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 1.3 | 6.5 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 1.3 | 20.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 1.3 | 3.8 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) |
| 1.2 | 3.7 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 1.2 | 14.9 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 1.2 | 9.9 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 1.2 | 3.7 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 1.2 | 4.8 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 1.2 | 22.9 | GO:0043383 | negative T cell selection(GO:0043383) |
| 1.2 | 4.8 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 1.1 | 13.4 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 1.1 | 3.3 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 1.1 | 8.8 | GO:0002568 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 1.1 | 6.6 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 1.1 | 3.2 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 1.1 | 4.3 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 1.1 | 3.2 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 1.1 | 10.6 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 1.0 | 4.2 | GO:0046722 | lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 1.0 | 4.9 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 1.0 | 6.9 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 1.0 | 4.8 | GO:0031296 | B cell costimulation(GO:0031296) |
| 1.0 | 4.8 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 1.0 | 2.9 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) regulation of Rap protein signal transduction(GO:0032487) negative regulation of integrin activation(GO:0033624) |
| 0.9 | 4.5 | GO:0009597 | detection of virus(GO:0009597) |
| 0.9 | 1.8 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.9 | 14.2 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.9 | 2.7 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.9 | 7.0 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.9 | 2.6 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) |
| 0.9 | 5.2 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.9 | 5.2 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.9 | 10.4 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.8 | 2.5 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.8 | 3.3 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.8 | 4.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.8 | 4.9 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.8 | 10.5 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) |
| 0.8 | 4.8 | GO:0032827 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.8 | 7.0 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.8 | 3.0 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.8 | 10.5 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.8 | 3.0 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.7 | 7.5 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.7 | 14.6 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.7 | 1.5 | GO:2000538 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.7 | 5.8 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.7 | 2.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.7 | 10.5 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.7 | 5.5 | GO:1903874 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) ferrous iron transmembrane transport(GO:1903874) |
| 0.7 | 4.0 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.7 | 3.9 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.7 | 2.0 | GO:1990051 | negative regulation of phospholipase C activity(GO:1900275) activation of protein kinase C activity(GO:1990051) |
| 0.7 | 9.8 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.7 | 6.5 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.7 | 13.7 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.6 | 1.3 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.6 | 3.9 | GO:0061017 | hepatoblast differentiation(GO:0061017) |
| 0.6 | 12.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.6 | 11.4 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.6 | 1.3 | GO:0002352 | B cell negative selection(GO:0002352) |
| 0.6 | 5.5 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.6 | 6.6 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.6 | 5.9 | GO:0032264 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.6 | 2.4 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.6 | 8.1 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.6 | 2.3 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.6 | 5.5 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.5 | 2.7 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.5 | 3.3 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.5 | 3.8 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.5 | 1.6 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.5 | 2.1 | GO:0051329 | interphase(GO:0051325) mitotic interphase(GO:0051329) |
| 0.5 | 6.9 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.5 | 1.6 | GO:0001810 | regulation of type I hypersensitivity(GO:0001810) positive regulation of type I hypersensitivity(GO:0001812) type I hypersensitivity(GO:0016068) |
| 0.5 | 8.9 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.5 | 6.8 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.5 | 6.6 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.5 | 1.5 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.5 | 2.0 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.5 | 1.5 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.5 | 3.0 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.5 | 1.0 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.5 | 32.1 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.5 | 2.9 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.5 | 2.9 | GO:0072539 | T-helper 17 cell differentiation(GO:0072539) |
| 0.5 | 3.9 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.5 | 3.9 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.5 | 1.4 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.5 | 1.4 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.5 | 6.2 | GO:1902563 | regulation of neutrophil degranulation(GO:0043313) regulation of neutrophil activation(GO:1902563) |
| 0.5 | 14.1 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.5 | 85.8 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.5 | 1.8 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.5 | 0.9 | GO:0002604 | regulation of dendritic cell antigen processing and presentation(GO:0002604) positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.4 | 1.3 | GO:2000847 | negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.4 | 1.3 | GO:1903660 | regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.4 | 2.2 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.4 | 6.5 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.4 | 2.6 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.4 | 4.7 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.4 | 1.7 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.4 | 5.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.4 | 5.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.4 | 4.9 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.4 | 7.3 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.4 | 5.9 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.4 | 13.6 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.4 | 3.1 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.4 | 1.1 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.4 | 1.9 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.4 | 3.0 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.4 | 3.0 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.4 | 5.2 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.4 | 2.5 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.4 | 1.1 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.3 | 1.0 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.3 | 1.0 | GO:1904501 | negative regulation of mitotic cell cycle, embryonic(GO:0045976) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.3 | 5.7 | GO:0002820 | negative regulation of adaptive immune response(GO:0002820) |
| 0.3 | 2.6 | GO:0032733 | positive regulation of interleukin-10 production(GO:0032733) |
| 0.3 | 11.3 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.3 | 66.0 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.3 | 2.2 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.3 | 1.2 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.3 | 8.9 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.3 | 1.8 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.3 | 5.9 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.3 | 1.2 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.3 | 2.4 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.3 | 1.8 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.3 | 2.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.3 | 1.4 | GO:0001757 | somite specification(GO:0001757) |
| 0.3 | 0.3 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.3 | 1.4 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.3 | 4.8 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.3 | 1.9 | GO:0009313 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.3 | 1.9 | GO:0036005 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.3 | 1.1 | GO:0072023 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.3 | 1.6 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.3 | 14.9 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.3 | 2.4 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.3 | 2.6 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.3 | 2.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.3 | 0.5 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.3 | 3.0 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.3 | 5.3 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.2 | 1.2 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.2 | 2.7 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.2 | 1.7 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.2 | 1.2 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.2 | 9.8 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.2 | 0.7 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.2 | 2.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.2 | 21.2 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.2 | 1.9 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.2 | 0.7 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.2 | 1.6 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.2 | 3.2 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.2 | 3.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.2 | 18.5 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.2 | 1.4 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.2 | 1.8 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.2 | 0.9 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.2 | 1.3 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.2 | 2.1 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.2 | 0.6 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.2 | 1.5 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.2 | 0.6 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.2 | 1.5 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.2 | 1.9 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.2 | 2.7 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) |
| 0.2 | 1.2 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.2 | 1.6 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.2 | 1.2 | GO:1900245 | negative regulation of MDA-5 signaling pathway(GO:0039534) positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.2 | 6.5 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.2 | 5.7 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.2 | 2.5 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.2 | 0.4 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.2 | 1.4 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.2 | 1.9 | GO:2000121 | regulation of removal of superoxide radicals(GO:2000121) |
| 0.2 | 1.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 5.0 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.2 | 1.5 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.2 | 1.5 | GO:0046638 | positive regulation of alpha-beta T cell differentiation(GO:0046638) |
| 0.2 | 2.4 | GO:0001553 | luteinization(GO:0001553) |
| 0.2 | 7.0 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.2 | 0.9 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.2 | 1.7 | GO:0014060 | regulation of epinephrine secretion(GO:0014060) negative regulation of epinephrine secretion(GO:0032811) |
| 0.2 | 2.0 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 | 0.7 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.2 | 0.9 | GO:0003418 | growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.2 | 1.4 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.2 | 0.7 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.2 | 1.0 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.2 | 1.9 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.2 | 2.5 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.2 | 0.7 | GO:0002191 | formation of cytoplasmic translation initiation complex(GO:0001732) cap-dependent translational initiation(GO:0002191) |
| 0.2 | 3.4 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.2 | 1.5 | GO:0050718 | positive regulation of interleukin-1 secretion(GO:0050716) positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.2 | 26.5 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.2 | 0.6 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.2 | 2.3 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.2 | 0.6 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.2 | 0.5 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 | 0.9 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 3.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 2.2 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.1 | 0.7 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 2.5 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 1.0 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.4 | GO:0046072 | dTDP biosynthetic process(GO:0006233) dTDP metabolic process(GO:0046072) |
| 0.1 | 8.6 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.1 | 0.6 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 0.3 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 | 0.7 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 0.6 | GO:0050812 | regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.1 | 1.9 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 1.0 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 3.4 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 2.6 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.1 | 9.6 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 0.7 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 4.6 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 0.8 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.1 | 2.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 0.5 | GO:1901301 | cochlear nucleus development(GO:0021747) regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 0.8 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.1 | 3.8 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 0.9 | GO:0071451 | removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) |
| 0.1 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.9 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.1 | 0.8 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.1 | 1.8 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 4.3 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 1.0 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 0.5 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.1 | 5.1 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 6.5 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 0.7 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.1 | 2.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 5.6 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.1 | 2.7 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 0.6 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.1 | 4.5 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 5.4 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 | 5.9 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 0.4 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 9.7 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 1.3 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.1 | 3.7 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 1.5 | GO:0072678 | T cell migration(GO:0072678) |
| 0.1 | 0.4 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 | 0.7 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.1 | 1.3 | GO:2000352 | negative regulation of endothelial cell apoptotic process(GO:2000352) |
| 0.1 | 4.6 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.1 | 0.4 | GO:0030730 | sequestering of triglyceride(GO:0030730) |
| 0.1 | 2.9 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.1 | 1.3 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 0.6 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.1 | 8.6 | GO:1903955 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 | 8.1 | GO:0030217 | T cell differentiation(GO:0030217) |
| 0.1 | 1.4 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 0.4 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.5 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 1.4 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.1 | 0.3 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 1.8 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.1 | 1.8 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.1 | 1.2 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 4.7 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 0.6 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.3 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.1 | 1.2 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.1 | 0.2 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 1.1 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 0.1 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.1 | 0.6 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.3 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.1 | 0.2 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 1.0 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.1 | 1.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 1.6 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 0.7 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 1.5 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.1 | 0.7 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.1 | 1.1 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.1 | 0.8 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.1 | 2.0 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 4.1 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 0.6 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 1.7 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.1 | 0.9 | GO:1901739 | regulation of myoblast fusion(GO:1901739) positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.2 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.1 | 0.6 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 0.2 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 0.1 | 1.0 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.1 | 0.3 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.4 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.6 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 1.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 0.4 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.8 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.4 | GO:0002097 | tRNA wobble base modification(GO:0002097) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 3.1 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.2 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.0 | 1.5 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 4.0 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 15.7 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 2.1 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.4 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 1.2 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 | 1.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 3.4 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 0.5 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) response to increased oxygen levels(GO:0036296) |
| 0.0 | 1.2 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 5.1 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.3 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.4 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.0 | 2.1 | GO:1901799 | negative regulation of proteasomal protein catabolic process(GO:1901799) |
| 0.0 | 0.3 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 | 1.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.4 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.3 | GO:0035909 | aorta morphogenesis(GO:0035909) |
| 0.0 | 0.6 | GO:1901099 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.2 | GO:0097324 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 1.1 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 1.4 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.7 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 0.2 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 1.0 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 0.7 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 3.8 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.4 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.1 | GO:1903299 | regulation of glucokinase activity(GO:0033131) regulation of hexokinase activity(GO:1903299) |
| 0.0 | 1.2 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 0.0 | 0.6 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 2.4 | GO:0007051 | spindle organization(GO:0007051) |
| 0.0 | 0.6 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.1 | GO:0061187 | histone H4-K20 demethylation(GO:0035574) regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.9 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.0 | 0.1 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.9 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 5.1 | GO:0034097 | response to cytokine(GO:0034097) |
| 0.0 | 0.1 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.3 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.5 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 22.3 | 111.5 | GO:0070442 | integrin alphaIIb-beta3 complex(GO:0070442) |
| 9.0 | 45.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 4.3 | 21.6 | GO:0044194 | cytolytic granule(GO:0044194) |
| 3.9 | 39.4 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 3.8 | 64.5 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 3.6 | 10.9 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 2.7 | 13.6 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 2.5 | 24.6 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 2.2 | 33.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 1.9 | 20.8 | GO:0042629 | mast cell granule(GO:0042629) |
| 1.8 | 5.3 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 1.7 | 6.9 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 1.6 | 19.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 1.6 | 22.3 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 1.5 | 3.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 1.5 | 4.6 | GO:0032997 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 1.5 | 1.5 | GO:1990423 | RZZ complex(GO:1990423) |
| 1.4 | 15.0 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 1.3 | 7.9 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 1.3 | 6.5 | GO:0097059 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 1.3 | 3.9 | GO:0060187 | cell pole(GO:0060187) |
| 1.2 | 14.9 | GO:0032009 | early phagosome(GO:0032009) |
| 1.2 | 4.8 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 1.1 | 5.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 1.1 | 5.4 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 1.1 | 10.6 | GO:1990462 | omegasome(GO:1990462) |
| 0.8 | 6.5 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.8 | 4.8 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.8 | 8.6 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.7 | 8.8 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.7 | 2.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.7 | 9.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.7 | 2.6 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.6 | 3.8 | GO:0032133 | chromosome passenger complex(GO:0032133) germinal vesicle(GO:0042585) |
| 0.6 | 6.9 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.6 | 10.6 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.6 | 4.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.5 | 0.5 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.5 | 4.7 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.5 | 27.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.5 | 1.5 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.5 | 2.8 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.5 | 1.4 | GO:0070877 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.5 | 7.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.4 | 11.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.4 | 5.9 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.4 | 1.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.4 | 135.3 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.4 | 4.5 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.4 | 1.6 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.4 | 223.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.4 | 1.6 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.4 | 4.0 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.4 | 1.9 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.4 | 7.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.3 | 4.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.3 | 9.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.3 | 19.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.3 | 1.9 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.3 | 1.9 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.3 | 1.5 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.3 | 9.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.3 | 11.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 0.7 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.2 | 11.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.2 | 14.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 1.5 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.2 | 6.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 0.8 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.2 | 2.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 4.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.2 | 16.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.2 | 0.6 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.2 | 2.9 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.2 | 7.5 | GO:0097546 | ciliary base(GO:0097546) |
| 0.2 | 1.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.2 | 18.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.2 | 2.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 2.8 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 2.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 1.0 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 6.6 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 0.8 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 1.9 | GO:0044217 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.1 | 25.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 1.5 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.1 | 1.6 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 2.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 0.3 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 0.3 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 1.3 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 1.0 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 11.1 | GO:0005930 | axoneme(GO:0005930) |
| 0.1 | 0.9 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 2.2 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 1.1 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 0.8 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.2 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.1 | 2.3 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 7.5 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 10.0 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 0.6 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 1.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.4 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 1.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 2.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.7 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 1.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 2.5 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.1 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 2.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 1.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.9 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 7.7 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.7 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.4 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 3.9 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 5.3 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 1.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 3.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 5.7 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 2.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 2.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.5 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.9 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 2.5 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 2.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 1.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.7 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 3.2 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 4.0 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 4.7 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 0.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.9 | 54.6 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 7.1 | 28.4 | GO:0031720 | haptoglobin binding(GO:0031720) hemoglobin alpha binding(GO:0031721) |
| 5.6 | 16.7 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 4.6 | 13.8 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 3.2 | 25.4 | GO:0019863 | IgE binding(GO:0019863) |
| 2.8 | 25.4 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 2.7 | 13.6 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 2.6 | 7.9 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 2.4 | 14.6 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 2.4 | 9.7 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 2.3 | 7.0 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 2.2 | 6.6 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 2.2 | 10.9 | GO:0035478 | chylomicron binding(GO:0035478) |
| 2.0 | 16.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 2.0 | 5.9 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 1.9 | 5.8 | GO:0042602 | riboflavin reductase (NADPH) activity(GO:0042602) |
| 1.9 | 5.6 | GO:0036004 | GAF domain binding(GO:0036004) |
| 1.7 | 15.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 1.6 | 8.2 | GO:0004131 | cytosine deaminase activity(GO:0004131) |
| 1.5 | 11.8 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 1.3 | 26.8 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 1.3 | 9.3 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 1.3 | 3.9 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 1.3 | 5.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 1.2 | 3.7 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 1.2 | 11.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 1.2 | 10.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 1.1 | 6.6 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 1.1 | 4.4 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 1.1 | 3.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 1.0 | 14.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 1.0 | 6.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 1.0 | 28.1 | GO:0008009 | chemokine activity(GO:0008009) |
| 1.0 | 19.7 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 1.0 | 4.9 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.9 | 2.7 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.9 | 5.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.9 | 10.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.8 | 6.8 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.8 | 5.9 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.8 | 76.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.8 | 17.4 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.8 | 1.7 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.8 | 2.4 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.8 | 5.5 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.8 | 18.7 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.8 | 10.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.8 | 3.9 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.8 | 23.8 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.8 | 4.5 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.7 | 3.6 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.7 | 1.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.7 | 13.7 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.7 | 9.9 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.7 | 2.0 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.7 | 2.6 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.6 | 1.9 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.6 | 1.3 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.6 | 2.6 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.6 | 5.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.6 | 5.9 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.6 | 2.4 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.6 | 2.3 | GO:0051435 | BH4 domain binding(GO:0051435) |
| 0.6 | 3.9 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.5 | 3.3 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.5 | 6.5 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.5 | 4.8 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.5 | 12.2 | GO:0050664 | oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.5 | 1.6 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.5 | 3.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.5 | 6.5 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.5 | 2.0 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.5 | 3.4 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.5 | 2.4 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.5 | 5.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.5 | 2.9 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.5 | 8.1 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.5 | 1.4 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.5 | 1.4 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.5 | 11.3 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.4 | 3.0 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.4 | 53.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.4 | 13.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.4 | 7.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.4 | 7.4 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.4 | 1.9 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.4 | 1.2 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.4 | 1.1 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.4 | 3.4 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.4 | 4.9 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.4 | 12.5 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.4 | 21.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.4 | 1.4 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.4 | 4.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.3 | 18.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.3 | 59.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.3 | 5.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.3 | 5.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.3 | 3.3 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.3 | 1.6 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.3 | 1.2 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.3 | 1.9 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.3 | 7.7 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.3 | 1.8 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.3 | 29.2 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.3 | 13.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.3 | 1.2 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.3 | 1.7 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.3 | 4.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.3 | 8.5 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.3 | 3.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.3 | 3.0 | GO:0043426 | MRF binding(GO:0043426) |
| 0.3 | 0.8 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.3 | 1.9 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.3 | 14.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.3 | 26.0 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.3 | 16.4 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.3 | 1.5 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.3 | 1.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 6.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.2 | 0.7 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.2 | 4.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 2.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.2 | 1.7 | GO:0070404 | NADH binding(GO:0070404) |
| 0.2 | 10.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.2 | 1.4 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.2 | 9.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.2 | 3.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 5.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 1.4 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.2 | 1.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.2 | 2.9 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.2 | 0.7 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.2 | 0.6 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 0.2 | 1.5 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.2 | 36.7 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.2 | 1.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.2 | 5.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.2 | 4.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 6.9 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.2 | 0.7 | GO:0038100 | nodal binding(GO:0038100) |
| 0.2 | 3.6 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 1.0 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.2 | 7.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 15.5 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 0.9 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.4 | GO:0004798 | thymidylate kinase activity(GO:0004798) |
| 0.1 | 0.6 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 16.5 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 5.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 1.1 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 29.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 2.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.6 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.1 | 1.9 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 2.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 4.5 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 2.0 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 2.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 1.0 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 3.6 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.1 | 3.7 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 1.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 2.8 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 1.5 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 1.9 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.1 | 2.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.3 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.1 | 1.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 1.9 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 0.5 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.1 | 1.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 1.4 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 0.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 3.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 5.1 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 4.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 3.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 2.6 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 2.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.5 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 7.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.7 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.9 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.9 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 1.4 | GO:0042287 | MHC protein binding(GO:0042287) MHC class I protein binding(GO:0042288) |
| 0.1 | 0.5 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.1 | 4.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.6 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 0.2 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 24.2 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 2.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.4 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.5 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 2.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 3.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.1 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.1 | 1.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.6 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 4.2 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 9.4 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 0.7 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.4 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 0.8 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 7.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 0.9 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 3.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.4 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 8.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.8 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 0.6 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 7.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.3 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.3 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 0.3 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.7 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.8 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.8 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.5 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 9.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 5.4 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 3.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 1.2 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.8 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.1 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 1.4 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 13.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.3 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 1.4 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.2 | GO:0031779 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 3.9 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 4.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 2.5 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.5 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 2.2 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 1.4 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.4 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 1.1 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.6 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.0 | GO:0001003 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 32.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 1.4 | 113.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 1.0 | 108.2 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 1.0 | 57.8 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.8 | 16.7 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.7 | 7.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.7 | 33.5 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.6 | 15.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.5 | 22.9 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.5 | 23.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.5 | 24.3 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.4 | 20.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.4 | 1.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.4 | 15.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.4 | 5.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.4 | 11.2 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.4 | 45.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.4 | 10.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.4 | 31.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.4 | 5.8 | PID EPO PATHWAY | EPO signaling pathway |
| 0.3 | 13.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.3 | 7.0 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.3 | 6.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 2.8 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.2 | 13.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.2 | 14.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 7.0 | ST ADRENERGIC | Adrenergic Pathway |
| 0.2 | 4.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 16.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.2 | 2.0 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.2 | 4.0 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.2 | 11.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.2 | 5.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.2 | 5.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.2 | 12.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 14.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 6.3 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 3.3 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.1 | 5.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 3.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 4.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 1.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 14.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 4.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 2.6 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 1.7 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 0.5 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 0.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 1.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.3 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 1.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 2.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.0 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 2.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 4.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 2.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 98.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 1.9 | 3.8 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 1.7 | 41.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 1.4 | 76.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 1.3 | 19.7 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 1.1 | 18.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 1.1 | 10.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 1.0 | 5.0 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 1.0 | 46.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 1.0 | 51.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 1.0 | 4.8 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.9 | 15.8 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.8 | 26.5 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.8 | 18.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.7 | 8.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.6 | 9.0 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.4 | 12.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.4 | 22.4 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.4 | 21.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.4 | 13.1 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.3 | 9.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.3 | 8.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.3 | 3.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 4.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.3 | 4.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.3 | 6.0 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.3 | 9.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.3 | 10.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.3 | 3.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.3 | 20.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.3 | 2.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.3 | 13.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.2 | 6.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 10.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.2 | 5.5 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.2 | 13.0 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.2 | 13.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.2 | 2.8 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 8.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 2.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 1.8 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.2 | 3.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.2 | 2.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 2.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 4.0 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 5.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.2 | 2.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.2 | 2.4 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 4.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 0.7 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 2.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 13.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 9.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 1.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 28.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.2 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.1 | 8.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 3.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 24.5 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 0.9 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.1 | 1.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 3.0 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.5 | REACTOME G PROTEIN BETA GAMMA SIGNALLING | Genes involved in G-protein beta:gamma signalling |
| 0.1 | 8.6 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.1 | 1.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 3.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 4.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 0.5 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.1 | 3.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 2.1 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.1 | 3.8 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 2.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 0.3 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.1 | 0.4 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 6.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 9.5 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.1 | 0.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 0.8 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 2.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.3 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 2.7 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.1 | 2.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 1.2 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.1 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 2.4 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 2.3 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.3 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.4 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 1.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.8 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.4 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |