Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Runx3
Z-value: 0.86
Transcription factors associated with Runx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Runx3
|
ENSMUSG00000070691.11 | Runx3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Runx3 | mm39_v1_chr4_+_134847949_134847973 | -0.23 | 5.2e-02 | Click! |
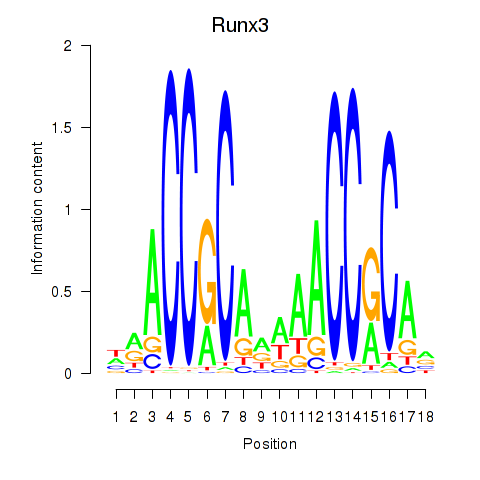
Activity profile of Runx3 motif
Sorted Z-values of Runx3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Runx3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_78739717 | 16.40 |
ENSMUST00000044584.6
|
Lgals2
|
lectin, galactose-binding, soluble 2 |
| chr16_+_4825216 | 9.97 |
ENSMUST00000185147.8
|
Smim22
|
small integral membrane protein 22 |
| chr7_-_25516041 | 9.07 |
ENSMUST00000043314.10
|
Cyp2s1
|
cytochrome P450, family 2, subfamily s, polypeptide 1 |
| chr11_-_100098333 | 7.03 |
ENSMUST00000007272.8
|
Krt14
|
keratin 14 |
| chr16_-_20972750 | 5.66 |
ENSMUST00000170665.3
|
Teddm3
|
transmembrane epididymal family member 3 |
| chr7_+_140796537 | 5.38 |
ENSMUST00000141804.2
|
Rassf7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr14_-_20231871 | 4.92 |
ENSMUST00000024011.10
|
Kcnk5
|
potassium channel, subfamily K, member 5 |
| chr5_+_135382149 | 4.92 |
ENSMUST00000111180.9
ENSMUST00000065785.4 |
Trim50
|
tripartite motif-containing 50 |
| chr6_-_69626340 | 4.19 |
ENSMUST00000198328.2
|
Igkv4-53
|
immunoglobulin kappa variable 4-53 |
| chrX_+_98179725 | 4.08 |
ENSMUST00000052839.7
|
Efnb1
|
ephrin B1 |
| chr1_-_24139263 | 3.72 |
ENSMUST00000187369.7
ENSMUST00000187752.7 ENSMUST00000186999.7 |
Fam135a
|
family with sequence similarity 135, member A |
| chr1_-_24139387 | 3.26 |
ENSMUST00000027337.15
|
Fam135a
|
family with sequence similarity 135, member A |
| chr8_+_27513819 | 2.62 |
ENSMUST00000033873.9
ENSMUST00000211043.2 |
Erlin2
|
ER lipid raft associated 2 |
| chrX_-_146337046 | 2.51 |
ENSMUST00000112819.9
ENSMUST00000136789.8 |
Lrch2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr19_-_20368029 | 2.39 |
ENSMUST00000235280.2
|
Anxa1
|
annexin A1 |
| chr8_+_27513839 | 2.30 |
ENSMUST00000209563.2
ENSMUST00000209520.2 |
Erlin2
|
ER lipid raft associated 2 |
| chr5_+_45650821 | 2.28 |
ENSMUST00000198534.2
|
Lap3
|
leucine aminopeptidase 3 |
| chr4_-_133225849 | 2.26 |
ENSMUST00000125541.2
|
Trnp1
|
TMF1-regulated nuclear protein 1 |
| chr8_+_27513289 | 2.12 |
ENSMUST00000209795.2
ENSMUST00000209976.2 |
Erlin2
|
ER lipid raft associated 2 |
| chr10_+_76367427 | 1.95 |
ENSMUST00000048678.7
|
Lss
|
lanosterol synthase |
| chr5_+_45650716 | 1.93 |
ENSMUST00000046122.11
|
Lap3
|
leucine aminopeptidase 3 |
| chr10_-_24588030 | 1.88 |
ENSMUST00000105520.8
|
Enpp1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr16_-_10131804 | 1.74 |
ENSMUST00000078357.5
|
Emp2
|
epithelial membrane protein 2 |
| chr17_+_28547548 | 1.67 |
ENSMUST00000233895.2
ENSMUST00000232867.2 |
Rpl10a
|
ribosomal protein L10A |
| chr17_+_28547533 | 1.60 |
ENSMUST00000233427.2
ENSMUST00000233937.2 |
Rpl10a
|
ribosomal protein L10A |
| chr18_-_47501513 | 1.59 |
ENSMUST00000076043.13
ENSMUST00000135790.8 |
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr17_+_28547445 | 1.44 |
ENSMUST00000042334.16
|
Rpl10a
|
ribosomal protein L10A |
| chr15_+_98953776 | 1.36 |
ENSMUST00000229268.2
|
Prph
|
peripherin |
| chr14_-_72946972 | 1.26 |
ENSMUST00000162478.8
|
Fndc3a
|
fibronectin type III domain containing 3A |
| chr9_-_58065800 | 1.24 |
ENSMUST00000168864.4
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr10_+_79590910 | 1.23 |
ENSMUST00000219981.2
ENSMUST00000219228.2 ENSMUST00000020577.4 |
Fgf22
|
fibroblast growth factor 22 |
| chr13_+_46822992 | 1.18 |
ENSMUST00000099547.4
|
Fam8a1
|
family with sequence similarity 8, member A1 |
| chr5_+_33176160 | 1.16 |
ENSMUST00000019109.8
|
Ywhah
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide |
| chr2_+_145627900 | 1.08 |
ENSMUST00000110005.8
ENSMUST00000094480.11 |
Rin2
|
Ras and Rab interactor 2 |
| chr10_+_127919142 | 1.03 |
ENSMUST00000026459.6
|
Atp5b
|
ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit |
| chr7_+_140796559 | 0.99 |
ENSMUST00000148975.3
|
Rassf7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr9_-_78389006 | 0.97 |
ENSMUST00000042235.15
|
Eef1a1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr11_+_75570085 | 0.93 |
ENSMUST00000017920.14
ENSMUST00000108426.8 ENSMUST00000108425.8 ENSMUST00000093115.4 |
Crk
|
v-crk avian sarcoma virus CT10 oncogene homolog |
| chr13_+_51799268 | 0.89 |
ENSMUST00000075853.6
|
Cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr13_+_21919225 | 0.79 |
ENSMUST00000087714.6
|
H4c11
|
H4 clustered histone 11 |
| chr6_-_100648086 | 0.78 |
ENSMUST00000089245.7
ENSMUST00000113312.9 ENSMUST00000170667.8 |
Shq1
|
SHQ1 homolog (S. cerevisiae) |
| chr14_-_72947422 | 0.78 |
ENSMUST00000089017.12
|
Fndc3a
|
fibronectin type III domain containing 3A |
| chr11_-_5712357 | 0.69 |
ENSMUST00000120306.8
ENSMUST00000170116.8 ENSMUST00000053427.11 ENSMUST00000118076.8 |
Urgcp
|
upregulator of cell proliferation |
| chr13_-_115238427 | 0.68 |
ENSMUST00000224997.2
ENSMUST00000061673.9 |
Gm49395
Itga1
|
predicted gene, 49395 integrin alpha 1 |
| chr5_+_115149170 | 0.67 |
ENSMUST00000031530.9
|
Sppl3
|
signal peptide peptidase 3 |
| chr13_-_22016364 | 0.65 |
ENSMUST00000102979.2
|
H4c18
|
H4 clustered histone 18 |
| chr11_-_70111796 | 0.62 |
ENSMUST00000060010.3
ENSMUST00000190533.2 |
Slc16a13
|
solute carrier family 16 (monocarboxylic acid transporters), member 13 |
| chr11_+_86375441 | 0.60 |
ENSMUST00000020827.7
|
Rnft1
|
ring finger protein, transmembrane 1 |
| chr7_-_141794815 | 0.55 |
ENSMUST00000211591.2
|
Gm40460
|
predicted gene, 40460 |
| chr9_+_106306736 | 0.55 |
ENSMUST00000098994.7
ENSMUST00000059802.7 ENSMUST00000213448.2 ENSMUST00000217081.2 |
Rpl29
|
ribosomal protein L29 |
| chr11_-_70111602 | 0.52 |
ENSMUST00000141290.2
ENSMUST00000159867.2 |
Slc16a13
|
solute carrier family 16 (monocarboxylic acid transporters), member 13 |
| chr12_-_98225676 | 0.51 |
ENSMUST00000021390.9
|
Galc
|
galactosylceramidase |
| chr9_+_106306598 | 0.51 |
ENSMUST00000150576.8
|
Rpl29
|
ribosomal protein L29 |
| chr2_-_53081199 | 0.51 |
ENSMUST00000239398.2
ENSMUST00000076313.14 ENSMUST00000210789.3 |
Prpf40a
|
pre-mRNA processing factor 40A |
| chr1_+_58752415 | 0.47 |
ENSMUST00000114309.8
ENSMUST00000069333.8 |
Cflar
|
CASP8 and FADD-like apoptosis regulator |
| chr10_-_62438040 | 0.47 |
ENSMUST00000045866.9
|
Ddx21
|
DExD box helicase 21 |
| chr13_-_14237980 | 0.47 |
ENSMUST00000222687.2
ENSMUST00000221338.2 ENSMUST00000221713.2 ENSMUST00000170957.3 |
Ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr1_+_74317709 | 0.46 |
ENSMUST00000077985.4
|
Gpbar1
|
G protein-coupled bile acid receptor 1 |
| chr8_+_72662437 | 0.44 |
ENSMUST00000110002.8
ENSMUST00000125802.8 ENSMUST00000131544.8 |
Zfp882
Zfp617
|
zinc finger protein 882 zinc finger protein 617 |
| chr13_-_25204272 | 0.41 |
ENSMUST00000021772.4
|
Mrs2
|
MRS2 magnesium transporter |
| chr6_+_124690060 | 0.39 |
ENSMUST00000130279.2
|
Phb2
|
prohibitin 2 |
| chr11_-_57722830 | 0.37 |
ENSMUST00000036917.3
|
Hand1
|
heart and neural crest derivatives expressed 1 |
| chr16_-_94327689 | 0.37 |
ENSMUST00000023615.7
|
Vps26c
|
VPS26 endosomal protein sorting factor C |
| chr13_-_14237958 | 0.37 |
ENSMUST00000223174.2
|
Ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr7_-_78432774 | 0.34 |
ENSMUST00000032841.7
|
Mrpl46
|
mitochondrial ribosomal protein L46 |
| chr10_+_42378193 | 0.33 |
ENSMUST00000105499.2
|
Snx3
|
sorting nexin 3 |
| chr13_+_38009951 | 0.27 |
ENSMUST00000138043.8
|
Rreb1
|
ras responsive element binding protein 1 |
| chr11_-_5828251 | 0.26 |
ENSMUST00000102922.10
|
Pold2
|
polymerase (DNA directed), delta 2, regulatory subunit |
| chr7_-_141017500 | 0.25 |
ENSMUST00000172654.8
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr9_-_107971640 | 0.24 |
ENSMUST00000081309.13
ENSMUST00000191985.2 |
Apeh
|
acylpeptide hydrolase |
| chr13_-_21934675 | 0.23 |
ENSMUST00000102983.2
|
H4c12
|
H4 clustered histone 12 |
| chr2_-_50186690 | 0.22 |
ENSMUST00000144143.8
ENSMUST00000102769.11 ENSMUST00000133768.2 |
Mmadhc
|
methylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria |
| chr10_-_116417333 | 0.22 |
ENSMUST00000218744.2
ENSMUST00000105267.8 ENSMUST00000105265.8 ENSMUST00000167706.8 ENSMUST00000168036.8 ENSMUST00000169921.8 ENSMUST00000020374.6 |
Cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr16_-_58647137 | 0.22 |
ENSMUST00000215069.2
ENSMUST00000079955.6 |
Olfr175
|
olfactory receptor 175 |
| chr13_+_14238361 | 0.21 |
ENSMUST00000129488.8
ENSMUST00000110536.8 ENSMUST00000110534.8 ENSMUST00000039538.15 ENSMUST00000110533.2 |
Arid4b
|
AT rich interactive domain 4B (RBP1-like) |
| chr5_+_21629932 | 0.21 |
ENSMUST00000056045.5
|
Fam185a
|
family with sequence similarity 185, member A |
| chr13_+_38010203 | 0.20 |
ENSMUST00000128570.9
|
Rreb1
|
ras responsive element binding protein 1 |
| chr1_-_83037090 | 0.19 |
ENSMUST00000193762.2
|
A030005K14Rik
|
RIKEN cDNA A030005K14 gene |
| chr7_-_141017742 | 0.18 |
ENSMUST00000019226.14
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chrX_-_95400846 | 0.18 |
ENSMUST00000179832.8
|
Hsf3
|
heat shock transcription factor 3 |
| chr16_+_92098166 | 0.18 |
ENSMUST00000063641.11
ENSMUST00000118064.2 |
Smim11
|
small integral membrane protein 11 |
| chr17_-_45744637 | 0.17 |
ENSMUST00000024727.10
|
Cdc5l
|
cell division cycle 5-like (S. pombe) |
| chr1_+_82891043 | 0.15 |
ENSMUST00000220768.2
|
A030005L19Rik
|
RIKEN cDNA A030005L19 gene |
| chrX_-_72502595 | 0.15 |
ENSMUST00000033737.15
ENSMUST00000077243.5 |
Haus7
|
HAUS augmin-like complex, subunit 7 |
| chr6_+_115908709 | 0.15 |
ENSMUST00000032471.9
|
Rho
|
rhodopsin |
| chr2_+_157756535 | 0.14 |
ENSMUST00000109523.2
|
Vstm2l
|
V-set and transmembrane domain containing 2-like |
| chrX_-_97934387 | 0.12 |
ENSMUST00000113826.8
ENSMUST00000033560.9 ENSMUST00000142267.2 |
Ophn1
|
oligophrenin 1 |
| chr7_-_141017525 | 0.10 |
ENSMUST00000106006.8
ENSMUST00000201710.4 |
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr14_-_98406977 | 0.08 |
ENSMUST00000071533.13
ENSMUST00000069334.8 |
Dach1
|
dachshund family transcription factor 1 |
| chr13_+_38009981 | 0.07 |
ENSMUST00000110238.10
|
Rreb1
|
ras responsive element binding protein 1 |
| chr3_-_7678785 | 0.03 |
ENSMUST00000194279.6
|
Il7
|
interleukin 7 |
| chr3_+_95566082 | 0.03 |
ENSMUST00000037947.15
ENSMUST00000178686.2 |
Mcl1
|
myeloid cell leukemia sequence 1 |
| chr3_-_7678796 | 0.02 |
ENSMUST00000192202.6
|
Il7
|
interleukin 7 |
| chr4_+_19575128 | 0.01 |
ENSMUST00000108253.8
ENSMUST00000029888.4 |
Rmdn1
|
regulator of microtubule dynamics 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 7.0 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.6 | 7.0 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.5 | 2.4 | GO:0097350 | neutrophil clearance(GO:0097350) |
| 0.4 | 1.2 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.3 | 0.9 | GO:1990859 | cellular response to endothelin(GO:1990859) |
| 0.3 | 9.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.3 | 0.8 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.2 | 4.9 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 1.9 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.2 | 1.7 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.1 | 1.0 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 | 4.7 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 0.4 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.1 | 0.5 | GO:1903943 | skeletal muscle atrophy(GO:0014732) negative regulation of myoblast fusion(GO:1901740) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 4.1 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.1 | 0.3 | GO:0070676 | intralumenal vesicle formation(GO:0070676) negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 | 0.5 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.7 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 2.2 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.4 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 1.6 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.5 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.2 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 2.3 | GO:0021696 | cerebellar cortex morphogenesis(GO:0021696) |
| 0.0 | 0.8 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.1 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 1.9 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 1.4 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 1.0 | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.0 | 0.7 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 5.7 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.5 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.3 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.0 | 4.2 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.9 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.5 | GO:2000810 | regulation of bicellular tight junction assembly(GO:2000810) |
| 0.0 | 1.1 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.1 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 1.2 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.2 | 0.7 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 0.2 | 2.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.2 | 1.4 | GO:0044299 | C-fiber(GO:0044299) |
| 0.1 | 6.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.0 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 0.7 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 4.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 1.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 4.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.5 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 2.3 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 13.8 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.5 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 4.2 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.3 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 1.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 1.2 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 2.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 16.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 1.0 | 7.0 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.4 | 1.9 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.3 | 2.4 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.3 | 9.1 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.3 | 1.0 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.2 | 4.9 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.2 | 0.8 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.9 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.7 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.5 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.1 | 7.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 5.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.7 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 4.2 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 1.9 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.0 | 0.5 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 1.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.5 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.5 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.2 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 1.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 5.9 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 1.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 4.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.1 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 19.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 4.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 0.9 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.7 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 1.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 9.1 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 2.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 6.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.0 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.2 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.9 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 1.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 2.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |