Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Rxra
Z-value: 1.01
Transcription factors associated with Rxra
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rxra
|
ENSMUSG00000015846.16 | Rxra |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rxra | mm39_v1_chr2_+_27566452_27566470 | 0.15 | 2.0e-01 | Click! |
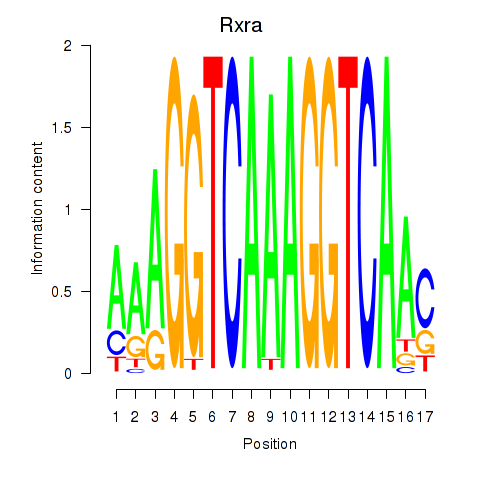
Activity profile of Rxra motif
Sorted Z-values of Rxra motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Rxra
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_78313043 | 12.06 |
ENSMUST00000001122.6
|
Slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr2_+_172994841 | 10.47 |
ENSMUST00000029017.6
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr11_-_53313950 | 7.04 |
ENSMUST00000036045.6
|
Leap2
|
liver-expressed antimicrobial peptide 2 |
| chr2_-_119060366 | 6.01 |
ENSMUST00000076084.6
|
Ppp1r14d
|
protein phosphatase 1, regulatory inhibitor subunit 14D |
| chr3_+_138121245 | 5.93 |
ENSMUST00000161312.8
ENSMUST00000013458.9 |
Adh4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr2_-_119060330 | 5.79 |
ENSMUST00000110820.3
|
Ppp1r14d
|
protein phosphatase 1, regulatory inhibitor subunit 14D |
| chr1_+_74448535 | 5.56 |
ENSMUST00000027366.13
|
Vil1
|
villin 1 |
| chr13_-_24098981 | 5.35 |
ENSMUST00000110407.4
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr7_-_19411866 | 5.11 |
ENSMUST00000142352.9
|
Apoc2
|
apolipoprotein C-II |
| chr12_-_103706774 | 5.07 |
ENSMUST00000186166.7
|
Serpina1b
|
serine (or cysteine) preptidase inhibitor, clade A, member 1B |
| chr13_-_24098951 | 5.06 |
ENSMUST00000021769.16
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr15_+_82336535 | 5.03 |
ENSMUST00000089129.7
ENSMUST00000229313.2 ENSMUST00000231136.2 |
Cyp2d9
|
cytochrome P450, family 2, subfamily d, polypeptide 9 |
| chr16_+_33614715 | 5.01 |
ENSMUST00000023520.7
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr12_-_103796632 | 4.96 |
ENSMUST00000164454.3
|
Serpina1b
|
serine (or cysteine) preptidase inhibitor, clade A, member 1B |
| chr11_-_116089866 | 4.79 |
ENSMUST00000066587.12
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr1_-_162641495 | 4.59 |
ENSMUST00000144916.8
ENSMUST00000140274.2 |
Fmo4
|
flavin containing monooxygenase 4 |
| chr16_+_33614378 | 4.56 |
ENSMUST00000115044.8
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr19_-_44017637 | 4.32 |
ENSMUST00000026211.10
ENSMUST00000211830.2 |
Cyp2c23
|
cytochrome P450, family 2, subfamily c, polypeptide 23 |
| chr18_+_50411431 | 3.90 |
ENSMUST00000039121.4
ENSMUST00000238078.2 |
Fam170a
|
family with sequence similarity 170, member A |
| chr10_+_62897353 | 3.83 |
ENSMUST00000178684.3
ENSMUST00000020266.15 |
Pbld1
|
phenazine biosynthesis-like protein domain containing 1 |
| chr19_-_21449916 | 3.58 |
ENSMUST00000087600.10
|
Gda
|
guanine deaminase |
| chr9_-_103165423 | 3.37 |
ENSMUST00000123530.8
|
1300017J02Rik
|
RIKEN cDNA 1300017J02 gene |
| chr6_+_58617521 | 3.37 |
ENSMUST00000145161.8
ENSMUST00000203146.3 ENSMUST00000114294.8 ENSMUST00000204948.2 |
Abcg2
|
ATP binding cassette subfamily G member 2 (Junior blood group) |
| chr9_-_103165489 | 3.35 |
ENSMUST00000035163.10
|
1300017J02Rik
|
RIKEN cDNA 1300017J02 gene |
| chr15_-_82264379 | 3.32 |
ENSMUST00000023083.9
|
Cyp2d22
|
cytochrome P450, family 2, subfamily d, polypeptide 22 |
| chr11_+_114566257 | 3.32 |
ENSMUST00000045779.6
|
Ttyh2
|
tweety family member 2 |
| chr3_-_144638284 | 3.25 |
ENSMUST00000098549.4
|
Clca4b
|
chloride channel accessory 4B |
| chr9_-_106448182 | 3.22 |
ENSMUST00000085111.5
|
Iqcf4
|
IQ motif containing F4 |
| chr6_+_78402956 | 3.05 |
ENSMUST00000079926.6
|
Reg1
|
regenerating islet-derived 1 |
| chr8_+_117822593 | 2.98 |
ENSMUST00000034308.16
ENSMUST00000176860.2 |
Bco1
|
beta-carotene oxygenase 1 |
| chr11_-_116089595 | 2.94 |
ENSMUST00000072948.11
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr5_+_92719336 | 2.87 |
ENSMUST00000176621.8
ENSMUST00000175974.2 ENSMUST00000131166.9 ENSMUST00000176448.8 ENSMUST00000082382.8 |
Fam47e
|
family with sequence similarity 47, member E |
| chr17_-_35351026 | 2.71 |
ENSMUST00000025249.7
|
Apom
|
apolipoprotein M |
| chr6_-_85797946 | 2.62 |
ENSMUST00000032074.5
|
Nat8f5
|
N-acetyltransferase 8 (GCN5-related) family member 5 |
| chr1_+_165957909 | 2.56 |
ENSMUST00000166159.2
|
Gpa33
|
glycoprotein A33 (transmembrane) |
| chr5_+_114142842 | 2.54 |
ENSMUST00000161610.6
|
Dao
|
D-amino acid oxidase |
| chr2_-_3420056 | 2.52 |
ENSMUST00000115084.8
ENSMUST00000115083.8 |
Meig1
|
meiosis expressed gene 1 |
| chr2_-_3420102 | 2.46 |
ENSMUST00000115082.10
|
Meig1
|
meiosis expressed gene 1 |
| chr19_+_39275518 | 2.41 |
ENSMUST00000003137.15
|
Cyp2c29
|
cytochrome P450, family 2, subfamily c, polypeptide 29 |
| chr15_-_77417512 | 2.40 |
ENSMUST00000062562.7
ENSMUST00000230863.2 |
Apol7c
|
apolipoprotein L 7c |
| chr2_-_121786573 | 2.38 |
ENSMUST00000104936.4
|
Mageb3
|
MAGE family member B3 |
| chr15_-_98155173 | 2.32 |
ENSMUST00000060855.7
|
H1f7
|
H1.7 linker histone |
| chr18_+_12732951 | 2.29 |
ENSMUST00000234255.2
ENSMUST00000169401.8 |
Ttc39c
|
tetratricopeptide repeat domain 39C |
| chr9_+_40059408 | 2.25 |
ENSMUST00000046333.9
ENSMUST00000238613.2 |
Tmem225
|
transmembrane protein 225 |
| chr5_+_114141894 | 2.20 |
ENSMUST00000086599.11
|
Dao
|
D-amino acid oxidase |
| chr16_+_4825216 | 2.19 |
ENSMUST00000185147.8
|
Smim22
|
small integral membrane protein 22 |
| chr14_+_69409251 | 2.18 |
ENSMUST00000062437.10
|
Nkx2-6
|
NK2 homeobox 6 |
| chr11_+_75422516 | 2.14 |
ENSMUST00000149727.8
ENSMUST00000108433.8 ENSMUST00000042561.14 ENSMUST00000143035.8 |
Slc43a2
|
solute carrier family 43, member 2 |
| chr16_+_4825170 | 2.14 |
ENSMUST00000178155.9
|
Smim22
|
small integral membrane protein 22 |
| chr19_+_36532061 | 2.13 |
ENSMUST00000169036.9
ENSMUST00000047247.12 |
Hectd2
|
HECT domain E3 ubiquitin protein ligase 2 |
| chr11_-_84761538 | 2.02 |
ENSMUST00000170741.2
ENSMUST00000172405.8 ENSMUST00000100686.10 ENSMUST00000108081.9 |
Ggnbp2
|
gametogenetin binding protein 2 |
| chr5_+_76988444 | 2.01 |
ENSMUST00000120639.9
ENSMUST00000163347.8 ENSMUST00000121851.2 |
Cracd
|
capping protein inhibiting regulator of actin |
| chr6_+_73225616 | 2.00 |
ENSMUST00000203632.2
|
Suclg1
|
succinate-CoA ligase, GDP-forming, alpha subunit |
| chr2_-_164041997 | 1.99 |
ENSMUST00000063251.3
|
Wfdc15a
|
WAP four-disulfide core domain 15A |
| chr16_+_4825146 | 1.91 |
ENSMUST00000184439.8
|
Smim22
|
small integral membrane protein 22 |
| chr19_+_38384428 | 1.91 |
ENSMUST00000054098.4
|
Slc35g1
|
solute carrier family 35, member G1 |
| chr1_-_172085977 | 1.90 |
ENSMUST00000111243.2
|
Atp1a4
|
ATPase, Na+/K+ transporting, alpha 4 polypeptide |
| chr19_+_5928649 | 1.87 |
ENSMUST00000136833.8
ENSMUST00000141362.2 |
Slc25a45
|
solute carrier family 25, member 45 |
| chr3_+_107784543 | 1.86 |
ENSMUST00000037375.10
ENSMUST00000199990.2 |
Eps8l3
|
EPS8-like 3 |
| chr5_-_31854942 | 1.82 |
ENSMUST00000031018.10
|
Rbks
|
ribokinase |
| chr2_-_32665596 | 1.80 |
ENSMUST00000161430.8
|
Ttc16
|
tetratricopeptide repeat domain 16 |
| chr8_+_88978600 | 1.80 |
ENSMUST00000154115.2
|
Tent4b
|
terminal nucleotidyltransferase 4B |
| chr2_+_164174660 | 1.78 |
ENSMUST00000017148.8
|
Svs5
|
seminal vesicle secretory protein 5 |
| chr5_-_87402659 | 1.77 |
ENSMUST00000075858.4
|
Ugt2b37
|
UDP glucuronosyltransferase 2 family, polypeptide B37 |
| chr10_+_14581325 | 1.75 |
ENSMUST00000191238.7
ENSMUST00000190114.2 |
Nmbr
|
neuromedin B receptor |
| chr1_+_139429430 | 1.71 |
ENSMUST00000027615.7
|
F13b
|
coagulation factor XIII, beta subunit |
| chr11_-_84761637 | 1.70 |
ENSMUST00000168434.8
|
Ggnbp2
|
gametogenetin binding protein 2 |
| chr19_-_6899121 | 1.69 |
ENSMUST00000173635.2
|
Esrra
|
estrogen related receptor, alpha |
| chr9_-_110818679 | 1.69 |
ENSMUST00000084922.6
ENSMUST00000199891.2 |
Rtp3
|
receptor transporter protein 3 |
| chr19_-_6899173 | 1.64 |
ENSMUST00000025906.12
ENSMUST00000239322.2 |
Esrra
|
estrogen related receptor, alpha |
| chr2_+_164158651 | 1.63 |
ENSMUST00000017144.3
|
Svs6
|
seminal vesicle secretory protein 6 |
| chr14_+_106343614 | 1.51 |
ENSMUST00000022708.7
|
Trim52
|
tripartite motif-containing 52 |
| chr7_-_103799746 | 1.46 |
ENSMUST00000059121.5
|
Ubqlnl
|
ubiquilin-like |
| chr13_-_49943717 | 1.45 |
ENSMUST00000220773.2
|
Gm30302
|
predicted gene, 30302 |
| chr15_-_97902576 | 1.45 |
ENSMUST00000023123.15
|
Col2a1
|
collagen, type II, alpha 1 |
| chr6_+_68495964 | 1.39 |
ENSMUST00000199510.5
ENSMUST00000103325.3 |
Igkv14-100
|
immunoglobulin kappa chain variable 14-100 |
| chr3_-_102871440 | 1.34 |
ENSMUST00000058899.13
|
Nr1h5
|
nuclear receptor subfamily 1, group H, member 5 |
| chr5_-_24760401 | 1.33 |
ENSMUST00000088302.10
|
Iqca1l
|
IQ motif containing with AAA domain 1 like |
| chr6_+_68279392 | 1.30 |
ENSMUST00000103322.3
|
Igkv2-109
|
immunoglobulin kappa variable 2-109 |
| chr15_-_97902515 | 1.26 |
ENSMUST00000088355.12
|
Col2a1
|
collagen, type II, alpha 1 |
| chr2_-_32665637 | 1.22 |
ENSMUST00000161958.2
|
Ttc16
|
tetratricopeptide repeat domain 16 |
| chr2_-_164435508 | 1.21 |
ENSMUST00000103100.2
|
Eppin
|
epididymal peptidase inhibitor |
| chr9_-_103105638 | 1.19 |
ENSMUST00000126359.2
|
Trf
|
transferrin |
| chr8_-_22550035 | 1.15 |
ENSMUST00000110738.3
|
Atp7b
|
ATPase, Cu++ transporting, beta polypeptide |
| chr12_-_113271532 | 1.14 |
ENSMUST00000192188.3
ENSMUST00000103418.3 |
Ighg2b
|
immunoglobulin heavy constant gamma 2B |
| chr16_+_33338898 | 1.12 |
ENSMUST00000119173.8
|
Slc12a8
|
solute carrier family 12 (potassium/chloride transporters), member 8 |
| chr1_+_190769010 | 1.11 |
ENSMUST00000077889.8
|
Spata45
|
spermatogenesis associated 45 |
| chr16_+_33338648 | 1.10 |
ENSMUST00000122427.8
ENSMUST00000059056.15 |
Slc12a8
|
solute carrier family 12 (potassium/chloride transporters), member 8 |
| chr2_-_19558719 | 1.09 |
ENSMUST00000062060.5
|
4921504E06Rik
|
RIKEN cDNA 4921504E06 gene |
| chr3_-_153610368 | 1.08 |
ENSMUST00000188338.7
|
Msh4
|
mutS homolog 4 |
| chr3_-_153610528 | 1.08 |
ENSMUST00000190449.2
|
Msh4
|
mutS homolog 4 |
| chr11_-_69906171 | 1.06 |
ENSMUST00000018718.8
ENSMUST00000102574.10 |
Acadvl
|
acyl-Coenzyme A dehydrogenase, very long chain |
| chr2_+_155453103 | 1.06 |
ENSMUST00000092995.6
|
Myh7b
|
myosin, heavy chain 7B, cardiac muscle, beta |
| chr2_-_32665342 | 1.06 |
ENSMUST00000161089.8
ENSMUST00000066478.9 ENSMUST00000161950.8 ENSMUST00000091059.12 |
Ttc16
|
tetratricopeptide repeat domain 16 |
| chr11_+_5578738 | 1.05 |
ENSMUST00000137933.2
|
Ankrd36
|
ankyrin repeat domain 36 |
| chr19_+_20470056 | 1.05 |
ENSMUST00000225337.3
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr16_-_64299511 | 1.04 |
ENSMUST00000089279.5
|
Csnka2ip
|
casein kinase 2, alpha prime interacting protein |
| chr11_+_109376432 | 1.02 |
ENSMUST00000106697.8
|
Arsg
|
arylsulfatase G |
| chr2_-_69172944 | 1.01 |
ENSMUST00000102709.8
ENSMUST00000102710.10 ENSMUST00000180142.2 |
Abcb11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr9_+_108270020 | 1.00 |
ENSMUST00000035234.6
|
1700102P08Rik
|
RIKEN cDNA 1700102P08 gene |
| chr7_-_43906629 | 1.00 |
ENSMUST00000012921.9
|
Acp4
|
acid phosphatase 4 |
| chr6_+_124689241 | 0.99 |
ENSMUST00000004375.16
|
Phb2
|
prohibitin 2 |
| chr2_+_23097482 | 0.99 |
ENSMUST00000028113.10
ENSMUST00000114505.2 |
Potegl
|
POTE ankyrin domain family, member G like |
| chrX_+_36763005 | 0.97 |
ENSMUST00000115172.12
ENSMUST00000185008.8 ENSMUST00000183696.2 |
Rhox3f
|
reproductive homeobox 3F |
| chr17_-_80885197 | 0.97 |
ENSMUST00000234602.2
|
Cdkl4
|
cyclin-dependent kinase-like 4 |
| chr5_-_24760427 | 0.96 |
ENSMUST00000198887.2
|
Iqca1l
|
IQ motif containing with AAA domain 1 like |
| chr7_-_97768689 | 0.94 |
ENSMUST00000138627.2
ENSMUST00000107127.8 |
Myo7a
|
myosin VIIA |
| chr14_-_14512458 | 0.93 |
ENSMUST00000211339.2
|
Gm45521
|
predicted gene 45521 |
| chrX_-_147398149 | 0.92 |
ENSMUST00000112755.3
|
Gm8334
|
predicted gene 8334 |
| chr14_+_73898665 | 0.90 |
ENSMUST00000098874.4
|
Gm21750
|
predicted gene, 21750 |
| chr18_+_70605476 | 0.90 |
ENSMUST00000114959.9
|
Stard6
|
StAR-related lipid transfer (START) domain containing 6 |
| chr11_-_11577824 | 0.89 |
ENSMUST00000081896.5
|
4930512M02Rik
|
RIKEN cDNA 4930512M02 gene |
| chr15_-_95426419 | 0.88 |
ENSMUST00000229933.2
ENSMUST00000166170.9 |
Nell2
|
NEL-like 2 |
| chr5_-_143831842 | 0.86 |
ENSMUST00000079624.12
ENSMUST00000110717.9 |
Ankrd61
|
ankyrin repeat domain 61 |
| chr2_+_155223728 | 0.85 |
ENSMUST00000043237.14
ENSMUST00000174685.8 |
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr9_+_108269992 | 0.84 |
ENSMUST00000192995.6
|
1700102P08Rik
|
RIKEN cDNA 1700102P08 gene |
| chr17_-_89508103 | 0.84 |
ENSMUST00000035701.6
|
Fshr
|
follicle stimulating hormone receptor |
| chr15_-_77331660 | 0.83 |
ENSMUST00000089469.7
|
Apol7b
|
apolipoprotein L 7b |
| chr15_-_100579813 | 0.80 |
ENSMUST00000230572.2
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr8_-_86107593 | 0.80 |
ENSMUST00000122452.8
|
Mylk3
|
myosin light chain kinase 3 |
| chr4_+_133795740 | 0.79 |
ENSMUST00000121391.8
|
Crybg2
|
crystallin beta-gamma domain containing 2 |
| chr19_-_9978987 | 0.79 |
ENSMUST00000117346.2
|
Best1
|
bestrophin 1 |
| chr16_+_29028860 | 0.79 |
ENSMUST00000162747.8
|
Plaat1
|
phospholipase A and acyltransferase 1 |
| chr7_-_28913382 | 0.78 |
ENSMUST00000169143.8
ENSMUST00000047846.13 |
Catsperg1
|
cation channel sperm associated auxiliary subunit gamma 1 |
| chrX_+_126301916 | 0.76 |
ENSMUST00000051530.4
|
4921511C20Rik
|
RIKEN cDNA 4921511C20 gene |
| chr15_-_77330396 | 0.75 |
ENSMUST00000229434.2
|
Apol7b
|
apolipoprotein L 7b |
| chr17_+_49922129 | 0.75 |
ENSMUST00000162854.2
|
Kif6
|
kinesin family member 6 |
| chr7_-_97768725 | 0.75 |
ENSMUST00000107128.8
|
Myo7a
|
myosin VIIA |
| chr9_+_38240356 | 0.74 |
ENSMUST00000214155.2
ENSMUST00000212354.3 |
Olfr25
|
olfactory receptor 25 |
| chr2_+_126057020 | 0.74 |
ENSMUST00000164042.3
|
Gm17555
|
predicted gene, 17555 |
| chr15_-_100579450 | 0.73 |
ENSMUST00000230740.2
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr12_-_115876396 | 0.72 |
ENSMUST00000103547.2
|
Ighv1-80
|
immunoglobulin heavy variable 1-80 |
| chr4_+_41569775 | 0.70 |
ENSMUST00000102963.10
|
Dnai1
|
dynein axonemal intermediate chain 1 |
| chr8_-_71085097 | 0.69 |
ENSMUST00000110103.2
|
Gdf15
|
growth differentiation factor 15 |
| chr7_-_19684654 | 0.68 |
ENSMUST00000043440.8
|
Igsf23
|
immunoglobulin superfamily, member 23 |
| chr13_-_24945844 | 0.68 |
ENSMUST00000006898.10
ENSMUST00000110382.9 |
Gmnn
|
geminin |
| chrX_+_36562041 | 0.67 |
ENSMUST00000096459.11
ENSMUST00000183543.8 ENSMUST00000183901.2 |
Rhox3a2
|
reproductive homeobox 3A2 |
| chrX_+_36518660 | 0.66 |
ENSMUST00000185028.8
ENSMUST00000185050.3 |
Rhox3a
|
reproductive homeobox 3A |
| chr4_-_103072343 | 0.66 |
ENSMUST00000150285.8
|
Slc35d1
|
solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1 |
| chr16_+_15135322 | 0.64 |
ENSMUST00000178312.2
|
Gm21897
|
predicted gene, 21897 |
| chrX_-_36843315 | 0.64 |
ENSMUST00000184824.7
ENSMUST00000185138.8 |
Rhox3h
|
reproductive homeobox 3H |
| chr9_-_48876290 | 0.62 |
ENSMUST00000008734.5
|
Htr3b
|
5-hydroxytryptamine (serotonin) receptor 3B |
| chr2_+_151841785 | 0.62 |
ENSMUST00000109861.8
ENSMUST00000109858.2 |
Slc52a3
|
solute carrier protein family 52, member 3 |
| chr1_-_74323536 | 0.60 |
ENSMUST00000190008.2
|
Aamp
|
angio-associated migratory protein |
| chr15_+_77583104 | 0.60 |
ENSMUST00000096358.6
|
Apol7e
|
apolipoprotein L 7e |
| chr2_+_167345009 | 0.58 |
ENSMUST00000078050.7
|
Rnf114
|
ring finger protein 114 |
| chr2_+_37029334 | 0.58 |
ENSMUST00000214905.2
ENSMUST00000217298.2 ENSMUST00000104995.3 |
Olfr364-ps1
|
olfactory receptor 364, pseudogene 1 |
| chr7_-_101513300 | 0.57 |
ENSMUST00000106981.8
|
Folr1
|
folate receptor 1 (adult) |
| chr8_-_100143029 | 0.56 |
ENSMUST00000155527.8
ENSMUST00000142129.8 ENSMUST00000093249.11 ENSMUST00000142475.3 ENSMUST00000128860.8 |
Cdh8
|
cadherin 8 |
| chr19_+_20470114 | 0.56 |
ENSMUST00000225313.2
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr14_+_122116002 | 0.56 |
ENSMUST00000039803.7
|
Ubac2
|
ubiquitin associated domain containing 2 |
| chr9_+_40098375 | 0.55 |
ENSMUST00000062229.6
|
Olfr986
|
olfactory receptor 986 |
| chrX_+_36651521 | 0.53 |
ENSMUST00000115189.10
ENSMUST00000123851.8 ENSMUST00000151387.2 |
Rhox3c
|
reproductive homeobox 3C |
| chr7_+_106413336 | 0.53 |
ENSMUST00000166880.3
ENSMUST00000075414.8 |
Olfr701
|
olfactory receptor 701 |
| chr9_+_44684248 | 0.52 |
ENSMUST00000128150.8
|
Ift46
|
intraflagellar transport 46 |
| chr12_+_57610897 | 0.51 |
ENSMUST00000101398.10
|
Ttc6
|
tetratricopeptide repeat domain 6 |
| chr11_-_53509485 | 0.48 |
ENSMUST00000000889.7
|
Il4
|
interleukin 4 |
| chr7_+_43093507 | 0.47 |
ENSMUST00000004729.5
ENSMUST00000206286.2 ENSMUST00000206196.2 ENSMUST00000206411.2 |
Etfb
|
electron transferring flavoprotein, beta polypeptide |
| chr10_+_23836392 | 0.47 |
ENSMUST00000092660.2
|
Taar4
|
trace amine-associated receptor 4 |
| chr9_-_108526548 | 0.46 |
ENSMUST00000013338.14
ENSMUST00000193197.6 |
Arih2
|
ariadne RBR E3 ubiquitin protein ligase 2 |
| chrX_-_36485911 | 0.45 |
ENSMUST00000115210.2
|
Rhox1
|
reproductive homeobox 1 |
| chr8_-_85741037 | 0.45 |
ENSMUST00000059072.6
ENSMUST00000209421.2 |
Best2
|
bestrophin 2 |
| chr2_-_21210151 | 0.45 |
ENSMUST00000027992.3
|
Enkur
|
enkurin, TRPC channel interacting protein |
| chr4_-_137301880 | 0.45 |
ENSMUST00000178923.2
|
Ldlrad2
|
low density lipoprotein receptor class A domain containing 2 |
| chr14_+_12016304 | 0.44 |
ENSMUST00000161302.8
|
Fhit
|
fragile histidine triad gene |
| chrX_+_36728628 | 0.44 |
ENSMUST00000081327.12
ENSMUST00000184210.8 ENSMUST00000184270.2 |
Rhox3e
|
reproductive homeobox 3E |
| chr12_-_74363168 | 0.43 |
ENSMUST00000110441.2
|
Gm11042
|
predicted gene 11042 |
| chr2_-_111100733 | 0.41 |
ENSMUST00000099619.6
|
Olfr1277
|
olfactory receptor 1277 |
| chr6_+_41498716 | 0.38 |
ENSMUST00000070380.5
|
Prss2
|
protease, serine 2 |
| chr17_-_28039588 | 0.38 |
ENSMUST00000114863.10
ENSMUST00000233131.2 |
Ilrun
|
inflammation and lipid regulator with UBA-like and NBR1-like domains |
| chr9_-_22046970 | 0.38 |
ENSMUST00000165735.9
|
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr5_+_20112771 | 0.38 |
ENSMUST00000200443.2
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chrX_-_36810265 | 0.38 |
ENSMUST00000119965.3
|
Rhox3g
|
reproductive homeobox 3G |
| chr11_+_50917831 | 0.38 |
ENSMUST00000072152.2
|
Olfr54
|
olfactory receptor 54 |
| chr10_+_74896383 | 0.38 |
ENSMUST00000164107.3
|
Bcr
|
BCR activator of RhoGEF and GTPase |
| chrX_-_88976458 | 0.37 |
ENSMUST00000088146.3
|
4930415L06Rik
|
RIKEN cDNA 4930415L06 gene |
| chrX_+_36561681 | 0.37 |
ENSMUST00000184565.8
|
Rhox3a2
|
reproductive homeobox 3A2 |
| chr11_+_4570067 | 0.36 |
ENSMUST00000109941.2
|
Gm11032
|
predicted gene 11032 |
| chrX_+_36762645 | 0.35 |
ENSMUST00000184451.8
|
Rhox3f
|
reproductive homeobox 3F |
| chr7_-_98305737 | 0.35 |
ENSMUST00000205911.2
ENSMUST00000038359.6 ENSMUST00000206611.2 ENSMUST00000206619.2 |
Emsy
|
EMSY, BRCA2-interacting transcriptional repressor |
| chr11_-_33463627 | 0.34 |
ENSMUST00000037522.14
|
Ranbp17
|
RAN binding protein 17 |
| chr17_+_6926452 | 0.32 |
ENSMUST00000097430.10
|
Sytl3
|
synaptotagmin-like 3 |
| chr11_-_97242842 | 0.32 |
ENSMUST00000093942.5
|
Gpr179
|
G protein-coupled receptor 179 |
| chr17_-_37422872 | 0.31 |
ENSMUST00000168659.2
|
Olfr92
|
olfactory receptor 92 |
| chr2_+_91033230 | 0.30 |
ENSMUST00000150403.8
ENSMUST00000002172.14 ENSMUST00000238832.2 ENSMUST00000239169.2 ENSMUST00000155418.2 |
Acp2
|
acid phosphatase 2, lysosomal |
| chr9_+_104424466 | 0.30 |
ENSMUST00000098443.9
|
Cpne4
|
copine IV |
| chr3_+_27371168 | 0.29 |
ENSMUST00000046383.12
|
Tnfsf10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr17_-_28039506 | 0.27 |
ENSMUST00000114859.9
ENSMUST00000233533.2 |
Ilrun
|
inflammation and lipid regulator with UBA-like and NBR1-like domains |
| chr1_-_74544946 | 0.27 |
ENSMUST00000044260.11
ENSMUST00000186282.7 |
Usp37
|
ubiquitin specific peptidase 37 |
| chr16_+_44632096 | 0.27 |
ENSMUST00000176819.8
ENSMUST00000176321.8 |
Cd200r4
|
CD200 receptor 4 |
| chr19_-_12302465 | 0.26 |
ENSMUST00000207241.3
|
Olfr1437
|
olfactory receptor 1437 |
| chr1_-_65158717 | 0.25 |
ENSMUST00000144760.3
|
Gm28845
|
predicted gene 28845 |
| chr13_+_33376931 | 0.25 |
ENSMUST00000067198.3
|
Serpinb9d
|
serine (or cysteine) peptidase inhibitor, clade B, member 9d |
| chr11_+_97689819 | 0.25 |
ENSMUST00000143571.2
|
Lasp1
|
LIM and SH3 protein 1 |
| chr9_+_44684450 | 0.25 |
ENSMUST00000238800.2
ENSMUST00000147559.8 |
Ift46
|
intraflagellar transport 46 |
| chr1_+_75456173 | 0.24 |
ENSMUST00000113575.9
ENSMUST00000148980.2 ENSMUST00000050899.7 ENSMUST00000187411.2 |
Tmem198
|
transmembrane protein 198 |
| chr8_-_4267459 | 0.24 |
ENSMUST00000176227.2
|
Prr36
|
proline rich 36 |
| chr6_+_8259379 | 0.24 |
ENSMUST00000162034.8
|
Umad1
|
UMAP1-MVP12 associated (UMA) domain containing 1 |
| chr13_+_33668761 | 0.23 |
ENSMUST00000081927.4
|
Serpinb9g
|
serine (or cysteine) peptidase inhibitor, clade B, member 9g |
| chr6_+_124690060 | 0.22 |
ENSMUST00000130279.2
|
Phb2
|
prohibitin 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 10.5 | GO:0006114 | glycerol biosynthetic process(GO:0006114) positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 1.6 | 4.7 | GO:0036088 | D-serine catabolic process(GO:0036088) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 1.5 | 5.9 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 1.4 | 5.6 | GO:1902896 | terminal web assembly(GO:1902896) |
| 1.3 | 7.7 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 1.3 | 5.1 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 1.3 | 3.8 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 1.1 | 3.4 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.9 | 2.7 | GO:0034443 | regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) |
| 0.9 | 3.6 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.6 | 9.6 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.6 | 4.6 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.5 | 2.2 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.5 | 2.8 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.5 | 1.8 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.4 | 3.3 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.4 | 1.1 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.3 | 0.8 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.3 | 1.9 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.3 | 3.2 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.3 | 1.0 | GO:0046618 | drug export(GO:0046618) |
| 0.2 | 1.5 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.2 | 2.7 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.2 | 0.6 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.2 | 1.2 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.2 | 2.0 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.2 | 3.0 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.2 | 1.6 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.2 | 1.2 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.2 | 1.1 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.2 | 0.5 | GO:2000422 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.2 | 10.0 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.2 | 2.3 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.1 | 2.2 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 0.8 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.1 | 1.7 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 1.1 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 | 0.5 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.1 | 0.8 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 5.5 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.1 | 0.9 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 24.4 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.1 | 3.3 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.1 | 0.7 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.6 | GO:0071231 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.1 | 14.3 | GO:0035304 | regulation of protein dephosphorylation(GO:0035304) |
| 0.1 | 0.4 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 3.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 0.6 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 3.3 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.4 | GO:0015961 | diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.0 | 2.6 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.0 | 0.4 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 1.7 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.0 | 0.1 | GO:0046166 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 2.1 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.0 | 0.9 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.3 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.4 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.7 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.2 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.4 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.3 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.5 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 6.8 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.4 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.6 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.5 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.6 | 1.7 | GO:1990427 | stereocilia tip link(GO:0002140) myosin VII complex(GO:0031477) stereocilia tip-link density(GO:1990427) upper tip-link density(GO:1990435) |
| 0.5 | 2.7 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.4 | 2.2 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.2 | 2.0 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.2 | 5.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.2 | 2.7 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 0.6 | GO:1904602 | serotonin-activated cation-selective channel complex(GO:1904602) |
| 0.1 | 12.5 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 0.5 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.1 | 1.2 | GO:0097433 | extrinsic component of external side of plasma membrane(GO:0031232) dense body(GO:0097433) |
| 0.1 | 3.0 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 0.5 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 4.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 1.0 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.8 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.8 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.7 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.6 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 13.0 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 4.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.9 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 1.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 17.9 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.5 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 2.0 | 5.9 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 1.2 | 4.7 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 1.1 | 7.7 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.9 | 5.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.7 | 5.6 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.7 | 2.0 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.7 | 10.4 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.6 | 1.8 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.5 | 2.7 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.4 | 2.4 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.4 | 4.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.4 | 3.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.4 | 1.1 | GO:0017099 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.3 | 1.6 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.3 | 1.1 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.2 | 11.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.2 | 0.6 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.2 | 2.2 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.2 | 1.7 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.2 | 1.2 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.2 | 2.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.2 | 5.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.2 | 1.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.6 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 0.5 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.1 | 0.4 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.1 | 3.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 1.7 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 0.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.7 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.7 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 1.0 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 12.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 2.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 2.1 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 1.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 12.2 | GO:0015293 | symporter activity(GO:0015293) |
| 0.1 | 1.0 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 1.4 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) |
| 0.1 | 0.6 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 4.6 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 1.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.8 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.7 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 2.0 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 3.3 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 2.7 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 3.7 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 1.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 6.5 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.0 | 1.0 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 1.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 2.3 | GO:0016853 | isomerase activity(GO:0016853) |
| 0.0 | 1.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 3.7 | GO:0005516 | calmodulin binding(GO:0005516) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 11.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 6.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 3.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 13.8 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.7 | 12.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.7 | 7.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.4 | 9.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.3 | 7.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.3 | 5.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.0 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 2.0 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 0.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 0.9 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 3.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 2.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 3.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.4 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.9 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 4.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 2.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.6 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |