Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
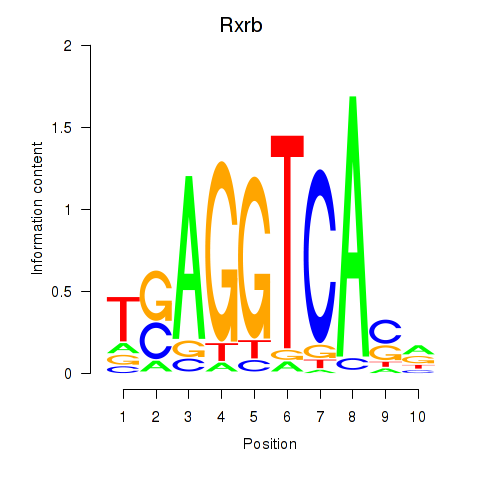
Results for Rxrb
Z-value: 1.09
Transcription factors associated with Rxrb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rxrb
|
ENSMUSG00000039656.18 | Rxrb |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rxrb | mm39_v1_chr17_+_34251322_34251381 | -0.19 | 1.0e-01 | Click! |
Activity profile of Rxrb motif
Sorted Z-values of Rxrb motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Rxrb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_38219447 | 16.35 |
ENSMUST00000081966.5
|
Odf1
|
outer dense fiber of sperm tails 1 |
| chr19_+_42040681 | 15.66 |
ENSMUST00000164518.4
|
4933411K16Rik
|
RIKEN cDNA 4933411K16 gene |
| chr2_+_154390808 | 12.82 |
ENSMUST00000045116.11
ENSMUST00000109709.4 |
1700003F12Rik
|
RIKEN cDNA 1700003F12 gene |
| chr1_-_156936197 | 12.50 |
ENSMUST00000187546.7
ENSMUST00000118207.8 ENSMUST00000027884.13 ENSMUST00000121911.8 |
Tex35
|
testis expressed 35 |
| chr7_-_4774277 | 12.40 |
ENSMUST00000174409.2
|
Fam71e2
|
family with sequence similarity 71, member E2 |
| chr1_-_190897012 | 11.16 |
ENSMUST00000171798.2
|
Fam71a
|
family with sequence similarity 71, member A |
| chr7_-_103778992 | 11.11 |
ENSMUST00000053743.6
|
Ubqln5
|
ubiquilin 5 |
| chr11_-_94376158 | 11.01 |
ENSMUST00000041705.8
|
Spata20
|
spermatogenesis associated 20 |
| chr8_+_12623016 | 10.54 |
ENSMUST00000210276.2
ENSMUST00000010579.8 ENSMUST00000209428.2 |
Spaca7
|
sperm acrosome associated 7 |
| chr7_+_45164005 | 10.54 |
ENSMUST00000210532.2
ENSMUST00000085331.14 ENSMUST00000210299.2 |
Tulp2
|
tubby-like protein 2 |
| chr2_-_153712996 | 9.84 |
ENSMUST00000028982.5
|
Sun5
|
Sad1 and UNC84 domain containing 5 |
| chr15_-_73702827 | 9.63 |
ENSMUST00000238402.2
|
Mroh5
|
maestro heat-like repeat family member 5 |
| chr6_+_90279064 | 9.60 |
ENSMUST00000167550.4
|
BC048671
|
cDNA sequence BC048671 |
| chr7_-_103792462 | 9.43 |
ENSMUST00000057254.6
|
Ubqln3
|
ubiquilin 3 |
| chr7_+_45164042 | 8.98 |
ENSMUST00000210868.2
|
Tulp2
|
tubby-like protein 2 |
| chr11_+_104077153 | 8.81 |
ENSMUST00000107000.2
ENSMUST00000059448.8 |
Sppl2c
|
signal peptide peptidase 2C |
| chr2_-_150934270 | 8.73 |
ENSMUST00000109890.2
|
Gm14151
|
predicted gene 14151 |
| chr8_+_110220614 | 8.72 |
ENSMUST00000034162.8
|
Pmfbp1
|
polyamine modulated factor 1 binding protein 1 |
| chr2_-_140231618 | 8.54 |
ENSMUST00000122367.8
ENSMUST00000120133.2 |
Sel1l2
|
sel-1 suppressor of lin-12-like 2 (C. elegans) |
| chr11_-_118233326 | 8.53 |
ENSMUST00000103024.4
|
Cep295nl
|
CEP295 N-terminal like |
| chr15_+_101345625 | 8.44 |
ENSMUST00000023781.9
|
Krt88
|
keratin 88 |
| chr1_+_175526152 | 8.35 |
ENSMUST00000094288.10
ENSMUST00000171939.8 |
Wdr64
|
WD repeat domain 64 |
| chr2_-_151318073 | 8.22 |
ENSMUST00000080132.3
|
4921509C19Rik
|
RIKEN cDNA 4921509C19 gene |
| chrX_-_100307592 | 8.19 |
ENSMUST00000101358.3
|
Gm614
|
predicted gene 614 |
| chr15_+_76152276 | 8.16 |
ENSMUST00000074173.4
|
Spatc1
|
spermatogenesis and centriole associated 1 |
| chr16_-_10608766 | 8.14 |
ENSMUST00000050864.7
|
Prm3
|
protamine 3 |
| chr1_+_85949657 | 7.95 |
ENSMUST00000052854.13
ENSMUST00000125083.2 ENSMUST00000152501.8 ENSMUST00000113344.8 ENSMUST00000130504.8 ENSMUST00000153247.3 |
Spata3
|
spermatogenesis associated 3 |
| chr4_-_63540653 | 7.35 |
ENSMUST00000102861.8
ENSMUST00000102862.4 |
Tex48
|
testis expressed 48 |
| chr1_+_133291302 | 7.33 |
ENSMUST00000135222.9
|
Etnk2
|
ethanolamine kinase 2 |
| chr11_-_23469181 | 7.28 |
ENSMUST00000239488.2
ENSMUST00000020527.13 |
1700093K21Rik
|
RIKEN cDNA 1700093K21 gene |
| chr11_+_101875095 | 7.18 |
ENSMUST00000176722.8
ENSMUST00000175972.2 |
Cfap97d1
|
CFAP97 domain containing 1 |
| chr2_-_25104628 | 7.13 |
ENSMUST00000043774.11
ENSMUST00000114363.2 |
Stpg3
|
sperm tail PG rich repeat containing 3 |
| chr2_+_150940165 | 7.05 |
ENSMUST00000109888.2
|
Gm14147
|
predicted gene 14147 |
| chr11_+_104983022 | 7.05 |
ENSMUST00000021029.6
|
Efcab3
|
EF-hand calcium binding domain 3 |
| chr1_+_85949681 | 6.91 |
ENSMUST00000159876.8
ENSMUST00000135440.2 |
Spata3
|
spermatogenesis associated 3 |
| chr4_-_106585127 | 6.83 |
ENSMUST00000106770.8
ENSMUST00000145044.2 |
Mroh7
|
maestro heat-like repeat family member 7 |
| chr14_-_21791544 | 6.78 |
ENSMUST00000184703.8
ENSMUST00000183698.8 ENSMUST00000119866.9 |
Dusp13
|
dual specificity phosphatase 13 |
| chr8_-_13612397 | 6.76 |
ENSMUST00000187391.7
ENSMUST00000134023.9 ENSMUST00000151400.10 |
1700029H14Rik
|
RIKEN cDNA 1700029H14 gene |
| chr11_-_3672188 | 6.57 |
ENSMUST00000102950.10
ENSMUST00000101632.4 |
Osbp2
|
oxysterol binding protein 2 |
| chr12_+_78243846 | 6.57 |
ENSMUST00000188791.2
|
Gm6657
|
predicted gene 6657 |
| chr1_+_84817547 | 6.34 |
ENSMUST00000097672.4
|
Fbxo36
|
F-box protein 36 |
| chr12_+_86781154 | 6.20 |
ENSMUST00000095527.6
|
Lrrc74a
|
leucine rich repeat containing 74A |
| chr5_-_137623331 | 6.18 |
ENSMUST00000031732.14
|
Fbxo24
|
F-box protein 24 |
| chr12_+_86781141 | 6.15 |
ENSMUST00000223308.2
|
Lrrc74a
|
leucine rich repeat containing 74A |
| chrX_+_85235370 | 6.15 |
ENSMUST00000026036.5
|
Nr0b1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr1_+_85949827 | 5.44 |
ENSMUST00000131151.3
|
Spata3
|
spermatogenesis associated 3 |
| chr14_-_118289557 | 5.34 |
ENSMUST00000022725.4
|
Dct
|
dopachrome tautomerase |
| chr7_-_119694400 | 5.16 |
ENSMUST00000209154.3
ENSMUST00000046993.4 |
Dnah3
|
dynein, axonemal, heavy chain 3 |
| chr4_-_117035922 | 5.09 |
ENSMUST00000153953.2
ENSMUST00000106436.8 |
Kif2c
|
kinesin family member 2C |
| chr12_-_111679344 | 5.02 |
ENSMUST00000160576.2
|
Bag5
|
BCL2-associated athanogene 5 |
| chr4_-_41048124 | 4.89 |
ENSMUST00000030136.13
|
Aqp7
|
aquaporin 7 |
| chr12_-_111679379 | 4.71 |
ENSMUST00000160825.2
ENSMUST00000162953.2 |
Bag5
|
BCL2-associated athanogene 5 |
| chr13_-_59892731 | 4.39 |
ENSMUST00000180139.3
|
1700014D04Rik
|
RIKEN cDNA 1700014D04 gene |
| chr12_-_87285478 | 4.35 |
ENSMUST00000222480.2
|
Noxred1
|
NADP+ dependent oxidoreductase domain containing 1 |
| chr5_+_124065481 | 4.25 |
ENSMUST00000166129.5
|
Gm43518
|
predicted gene 43518 |
| chr19_+_47926086 | 4.09 |
ENSMUST00000238163.2
ENSMUST00000066308.9 |
Cfap58
|
cilia and flagella associated protein 58 |
| chr2_-_79959802 | 4.00 |
ENSMUST00000102653.8
|
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr7_+_40636967 | 3.94 |
ENSMUST00000206529.2
ENSMUST00000171664.2 |
4930433I11Rik
|
RIKEN cDNA 4930433I11 gene |
| chr11_-_68864666 | 3.85 |
ENSMUST00000038644.5
|
Rangrf
|
RAN guanine nucleotide release factor |
| chr7_-_133966588 | 3.83 |
ENSMUST00000172947.8
|
D7Ertd443e
|
DNA segment, Chr 7, ERATO Doi 443, expressed |
| chr1_+_54369151 | 3.81 |
ENSMUST00000163072.8
|
Ccdc150
|
coiled-coil domain containing 150 |
| chr12_-_111679618 | 3.81 |
ENSMUST00000054636.7
|
Bag5
|
BCL2-associated athanogene 5 |
| chr9_-_40915858 | 3.45 |
ENSMUST00000188848.8
ENSMUST00000034519.13 |
Crtam
|
cytotoxic and regulatory T cell molecule |
| chr4_+_110254907 | 3.25 |
ENSMUST00000097920.9
ENSMUST00000080744.13 |
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr3_-_94693780 | 3.20 |
ENSMUST00000107273.9
ENSMUST00000238849.2 |
Cgn
|
cingulin |
| chr1_-_84817000 | 3.07 |
ENSMUST00000186648.7
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr17_+_35191661 | 3.02 |
ENSMUST00000007248.5
|
Hspa1l
|
heat shock protein 1-like |
| chr15_-_76193955 | 2.76 |
ENSMUST00000210024.2
|
Oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr1_-_84817022 | 2.71 |
ENSMUST00000189496.7
ENSMUST00000027421.13 ENSMUST00000186894.7 |
Trip12
|
thyroid hormone receptor interactor 12 |
| chr16_+_24540599 | 2.70 |
ENSMUST00000115314.4
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr9_+_45313913 | 2.63 |
ENSMUST00000214257.2
|
Fxyd2
|
FXYD domain-containing ion transport regulator 2 |
| chr5_+_73071607 | 2.46 |
ENSMUST00000144843.8
|
Slain2
|
SLAIN motif family, member 2 |
| chr6_-_116050081 | 2.42 |
ENSMUST00000173548.3
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr9_-_40915895 | 2.33 |
ENSMUST00000180384.3
|
Crtam
|
cytotoxic and regulatory T cell molecule |
| chr11_-_99501015 | 2.09 |
ENSMUST00000076478.2
|
Gm11937
|
predicted gene 11937 |
| chr2_-_79959178 | 2.09 |
ENSMUST00000102654.11
ENSMUST00000102655.10 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr6_+_129510331 | 2.08 |
ENSMUST00000204956.2
ENSMUST00000204639.2 |
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr12_+_91367764 | 2.08 |
ENSMUST00000021346.14
ENSMUST00000021343.8 |
Tshr
|
thyroid stimulating hormone receptor |
| chr4_+_110254858 | 2.04 |
ENSMUST00000106589.9
ENSMUST00000106587.9 ENSMUST00000106591.8 ENSMUST00000106592.8 |
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr16_-_23339548 | 2.01 |
ENSMUST00000089883.7
|
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr8_-_68363564 | 1.97 |
ENSMUST00000093468.12
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr6_-_124942170 | 1.97 |
ENSMUST00000148485.2
ENSMUST00000129976.8 |
Cops7a
|
COP9 signalosome subunit 7A |
| chr3_-_94693740 | 1.97 |
ENSMUST00000153263.9
ENSMUST00000107272.7 ENSMUST00000155485.4 |
Cgn
|
cingulin |
| chr5_+_73071897 | 1.92 |
ENSMUST00000200785.2
|
Slain2
|
SLAIN motif family, member 2 |
| chr11_-_99505668 | 1.91 |
ENSMUST00000074926.6
|
Krtap2-4
|
keratin associated protein 2-4 |
| chr11_+_100960838 | 1.80 |
ENSMUST00000001802.10
|
Naglu
|
alpha-N-acetylglucosaminidase (Sanfilippo disease IIIB) |
| chr2_+_48839505 | 1.71 |
ENSMUST00000112745.8
ENSMUST00000112754.8 |
Mbd5
|
methyl-CpG binding domain protein 5 |
| chr6_-_124942457 | 1.62 |
ENSMUST00000112439.9
|
Cops7a
|
COP9 signalosome subunit 7A |
| chr13_+_43276323 | 1.51 |
ENSMUST00000136576.8
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr16_-_23339329 | 1.50 |
ENSMUST00000230040.2
ENSMUST00000229619.2 |
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr1_-_38168697 | 1.50 |
ENSMUST00000027251.12
|
Rev1
|
REV1, DNA directed polymerase |
| chr19_-_37153436 | 1.49 |
ENSMUST00000142973.2
ENSMUST00000154376.8 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr7_+_137039309 | 1.49 |
ENSMUST00000064404.8
ENSMUST00000211496.2 ENSMUST00000209696.2 |
Glrx3
|
glutaredoxin 3 |
| chr7_-_105249308 | 1.49 |
ENSMUST00000210531.2
ENSMUST00000033185.10 |
Hpx
|
hemopexin |
| chr11_-_70146156 | 1.46 |
ENSMUST00000108574.3
ENSMUST00000000329.9 |
Alox12
|
arachidonate 12-lipoxygenase |
| chr10_-_87982732 | 1.45 |
ENSMUST00000164121.8
ENSMUST00000164803.2 ENSMUST00000168163.8 ENSMUST00000048518.16 |
Parpbp
|
PARP1 binding protein |
| chr6_-_124942366 | 1.44 |
ENSMUST00000129446.8
ENSMUST00000032220.15 |
Cops7a
|
COP9 signalosome subunit 7A |
| chr4_+_148085179 | 1.43 |
ENSMUST00000103230.5
|
Nppa
|
natriuretic peptide type A |
| chr15_-_101651534 | 1.42 |
ENSMUST00000023710.6
|
Krt71
|
keratin 71 |
| chr7_+_101879176 | 1.40 |
ENSMUST00000120119.9
|
Pgap2
|
post-GPI attachment to proteins 2 |
| chr6_-_88423464 | 1.37 |
ENSMUST00000204459.3
ENSMUST00000203213.3 ENSMUST00000205179.3 ENSMUST00000165242.4 |
Eefsec
|
eukaryotic elongation factor, selenocysteine-tRNA-specific |
| chr7_-_30755007 | 1.25 |
ENSMUST00000206474.2
ENSMUST00000205807.2 ENSMUST00000039909.13 ENSMUST00000206305.2 ENSMUST00000205439.2 |
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr5_+_73071695 | 1.25 |
ENSMUST00000143829.5
|
Slain2
|
SLAIN motif family, member 2 |
| chr17_+_25097199 | 1.23 |
ENSMUST00000050714.8
|
Igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr8_-_106732607 | 1.22 |
ENSMUST00000227778.2
|
Dpep2nb
|
Dpep2 neighbor |
| chr2_-_48839276 | 1.22 |
ENSMUST00000028098.11
|
Orc4
|
origin recognition complex, subunit 4 |
| chr10_-_128383508 | 1.21 |
ENSMUST00000152539.8
ENSMUST00000133458.2 ENSMUST00000040572.10 |
Zc3h10
|
zinc finger CCCH type containing 10 |
| chr19_+_11747721 | 1.20 |
ENSMUST00000167199.3
|
Mrpl16
|
mitochondrial ribosomal protein L16 |
| chr4_-_41275091 | 1.19 |
ENSMUST00000030143.13
ENSMUST00000108068.8 |
Ubap2
|
ubiquitin-associated protein 2 |
| chr1_+_133109059 | 1.12 |
ENSMUST00000187285.7
|
Plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr13_+_20274708 | 1.12 |
ENSMUST00000072519.7
|
Elmo1
|
engulfment and cell motility 1 |
| chr17_-_71575584 | 1.08 |
ENSMUST00000233148.2
|
Emilin2
|
elastin microfibril interfacer 2 |
| chr10_+_61556371 | 1.01 |
ENSMUST00000080099.6
|
Aifm2
|
apoptosis-inducing factor, mitochondrion-associated 2 |
| chr2_-_6217844 | 1.01 |
ENSMUST00000042658.5
|
Echdc3
|
enoyl Coenzyme A hydratase domain containing 3 |
| chr11_-_99511257 | 1.01 |
ENSMUST00000073853.3
|
Gm11562
|
predicted gene 11562 |
| chr17_-_75858835 | 1.00 |
ENSMUST00000234785.2
ENSMUST00000112507.4 |
Fam98a
|
family with sequence similarity 98, member A |
| chr6_+_125122172 | 0.99 |
ENSMUST00000119527.8
ENSMUST00000117675.8 |
Iffo1
|
intermediate filament family orphan 1 |
| chr5_+_137628377 | 0.95 |
ENSMUST00000175968.8
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr2_+_71884943 | 0.93 |
ENSMUST00000028525.6
|
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr18_+_35987733 | 0.92 |
ENSMUST00000235337.2
|
Cxxc5
|
CXXC finger 5 |
| chr10_+_79984097 | 0.90 |
ENSMUST00000099492.10
ENSMUST00000042057.12 |
Midn
|
midnolin |
| chr12_+_33364288 | 0.90 |
ENSMUST00000144586.2
|
Atxn7l1
|
ataxin 7-like 1 |
| chr6_+_129510117 | 0.80 |
ENSMUST00000032264.9
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chrX_-_99456185 | 0.77 |
ENSMUST00000033567.15
|
Awat2
|
acyl-CoA wax alcohol acyltransferase 2 |
| chr1_-_38168801 | 0.76 |
ENSMUST00000195383.2
|
Rev1
|
REV1, DNA directed polymerase |
| chr3_+_32791139 | 0.75 |
ENSMUST00000127477.8
ENSMUST00000121778.8 ENSMUST00000154257.8 |
Ndufb5
|
NADH:ubiquinone oxidoreductase subunit B5 |
| chr14_-_31552335 | 0.72 |
ENSMUST00000228037.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr6_-_124840824 | 0.71 |
ENSMUST00000046893.10
ENSMUST00000204667.2 |
Gpr162
|
G protein-coupled receptor 162 |
| chr2_-_48839218 | 0.68 |
ENSMUST00000090976.10
ENSMUST00000149679.8 |
Orc4
|
origin recognition complex, subunit 4 |
| chr1_-_52539395 | 0.66 |
ENSMUST00000186764.7
|
Nab1
|
Ngfi-A binding protein 1 |
| chr10_+_87982854 | 0.65 |
ENSMUST00000052355.15
|
Nup37
|
nucleoporin 37 |
| chr11_+_75084609 | 0.63 |
ENSMUST00000102514.4
|
Rtn4rl1
|
reticulon 4 receptor-like 1 |
| chr17_-_32607859 | 0.63 |
ENSMUST00000087703.12
ENSMUST00000170603.3 |
Wiz
|
widely-interspaced zinc finger motifs |
| chr7_+_105289655 | 0.62 |
ENSMUST00000058333.10
|
Timm10b
|
translocase of inner mitochondrial membrane 10B |
| chr13_+_73911797 | 0.62 |
ENSMUST00000017900.9
|
Slc12a7
|
solute carrier family 12, member 7 |
| chr4_-_45532470 | 0.60 |
ENSMUST00000147448.2
|
Shb
|
src homology 2 domain-containing transforming protein B |
| chr8_+_72860884 | 0.55 |
ENSMUST00000210435.4
|
Olfr374
|
olfactory receptor 374 |
| chr12_-_98703664 | 0.53 |
ENSMUST00000170188.8
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chrX_+_72760183 | 0.52 |
ENSMUST00000002084.14
|
Abcd1
|
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr3_+_96079642 | 0.52 |
ENSMUST00000076372.5
|
Sf3b4
|
splicing factor 3b, subunit 4 |
| chr10_-_128657445 | 0.43 |
ENSMUST00000217685.2
ENSMUST00000026409.5 ENSMUST00000219215.2 ENSMUST00000219524.2 |
Ormdl2
|
ORM1-like 2 (S. cerevisiae) |
| chr9_-_50571080 | 0.42 |
ENSMUST00000034567.4
|
Dlat
|
dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) |
| chr4_-_20778847 | 0.42 |
ENSMUST00000102998.4
|
Nkain3
|
Na+/K+ transporting ATPase interacting 3 |
| chr17_-_66756710 | 0.37 |
ENSMUST00000086693.12
ENSMUST00000097291.10 |
Mtcl1
|
microtubule crosslinking factor 1 |
| chr18_+_35987791 | 0.35 |
ENSMUST00000235404.2
|
Cxxc5
|
CXXC finger 5 |
| chr6_+_129510145 | 0.34 |
ENSMUST00000204487.3
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr15_-_101710781 | 0.33 |
ENSMUST00000063292.8
|
Krt73
|
keratin 73 |
| chr10_+_87982916 | 0.29 |
ENSMUST00000169309.3
|
Nup37
|
nucleoporin 37 |
| chr11_+_51541728 | 0.26 |
ENSMUST00000117859.8
ENSMUST00000064493.6 |
D930048N14Rik
|
RIKEN cDNA D930048N14 gene |
| chr11_+_117375194 | 0.23 |
ENSMUST00000092394.4
|
Gm11733
|
predicted gene 11733 |
| chr2_-_76478336 | 0.23 |
ENSMUST00000002808.7
|
Prkra
|
protein kinase, interferon inducible double stranded RNA dependent activator |
| chr10_+_127126643 | 0.22 |
ENSMUST00000026475.15
ENSMUST00000139091.2 ENSMUST00000230446.2 |
Ddit3
Ddit3
|
DNA-damage inducible transcript 3 DNA-damage inducible transcript 3 |
| chr5_-_25200745 | 0.22 |
ENSMUST00000076306.12
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr12_+_111679689 | 0.20 |
ENSMUST00000040519.12
ENSMUST00000163220.10 ENSMUST00000162316.2 |
Coa8
|
cytochrome c oxidase assembly factor 8 |
| chr2_+_118730823 | 0.19 |
ENSMUST00000151162.2
|
Bahd1
|
bromo adjacent homology domain containing 1 |
| chr14_+_54713703 | 0.17 |
ENSMUST00000164697.8
|
Rem2
|
rad and gem related GTP binding protein 2 |
| chr9_-_106769131 | 0.16 |
ENSMUST00000159283.8
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr10_-_100425067 | 0.15 |
ENSMUST00000218821.2
ENSMUST00000054471.10 |
4930430F08Rik
|
RIKEN cDNA 4930430F08 gene |
| chr15_-_101471324 | 0.11 |
ENSMUST00000023714.5
|
Krt90
|
keratin 90 |
| chr9_-_106769069 | 0.11 |
ENSMUST00000160503.4
ENSMUST00000159620.9 |
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr4_+_152262583 | 0.10 |
ENSMUST00000075363.10
|
Acot7
|
acyl-CoA thioesterase 7 |
| chr5_+_3853160 | 0.09 |
ENSMUST00000080085.9
ENSMUST00000200386.5 |
Krit1
|
KRIT1, ankyrin repeat containing |
| chr14_+_47710574 | 0.09 |
ENSMUST00000228740.2
|
Fbxo34
|
F-box protein 34 |
| chr4_-_58912678 | 0.08 |
ENSMUST00000144512.8
ENSMUST00000102889.10 ENSMUST00000055822.15 |
Ecpas
|
Ecm29 proteasome adaptor and scaffold |
| chr11_-_53321606 | 0.06 |
ENSMUST00000061326.5
ENSMUST00000109021.4 |
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr11_-_95037089 | 0.05 |
ENSMUST00000021241.8
|
Dlx4
|
distal-less homeobox 4 |
| chr16_+_31482745 | 0.05 |
ENSMUST00000100001.10
ENSMUST00000064477.14 |
Dlg1
|
discs large MAGUK scaffold protein 1 |
| chrX_-_7774074 | 0.03 |
ENSMUST00000085330.5
|
Gm10491
|
predicted gene 10491 |
| chr14_+_6226418 | 0.01 |
ENSMUST00000112625.9
|
Oxsm
|
3-oxoacyl-ACP synthase, mitochondrial |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 13.5 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 1.9 | 5.8 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 1.8 | 5.3 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 1.2 | 19.5 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 1.1 | 5.3 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 1.0 | 24.0 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 1.0 | 4.9 | GO:0015793 | glycerol transport(GO:0015793) renal water absorption(GO:0070295) |
| 1.0 | 5.8 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.8 | 5.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.7 | 7.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.7 | 6.2 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.7 | 9.8 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.6 | 10.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.5 | 4.3 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.5 | 1.5 | GO:1900247 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.5 | 2.3 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.4 | 3.9 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.4 | 1.7 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.4 | 1.5 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.4 | 1.4 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.3 | 5.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.3 | 2.1 | GO:1904587 | response to glycoprotein(GO:1904587) |
| 0.3 | 3.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.3 | 5.0 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.2 | 1.5 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) heme transport(GO:0015886) positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.2 | 3.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 0.9 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.2 | 8.1 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.2 | 6.1 | GO:0034391 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.1 | 1.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 1.4 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 3.8 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 1.4 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.8 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 5.6 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 1.2 | GO:1903797 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.1 | 0.9 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.2 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.1 | 0.4 | GO:1900060 | negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.1 | 3.0 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 0.3 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.1 | 0.6 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 1.8 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 5.2 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.1 | 1.0 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 0.2 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 | 0.6 | GO:1900194 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 2.0 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.4 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 33.9 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 1.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 1.9 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 1.4 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 2.6 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 2.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 2.4 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.6 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.4 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) positive regulation of microtubule motor activity(GO:2000576) |
| 0.0 | 0.1 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 1.1 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.6 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 1.2 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.7 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.7 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.0 | 4.9 | GO:0006470 | protein dephosphorylation(GO:0006470) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 8.8 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 1.2 | 16.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.7 | 24.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.4 | 5.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.4 | 6.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.3 | 8.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.2 | 0.9 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.2 | 1.9 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 9.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.2 | 8.1 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.2 | 7.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 3.0 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 0.4 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 5.3 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 1.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 13.5 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 3.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.6 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 1.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 5.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.4 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.9 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.2 | GO:1990622 | CHOP-C/EBP complex(GO:0036488) CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 1.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 10.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 1.7 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 6.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 36.2 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 1.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.4 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 5.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 2.0 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 6.9 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.2 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.2 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.0 | GO:0005811 | lipid particle(GO:0005811) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.3 | GO:0004103 | choline kinase activity(GO:0004103) |
| 1.1 | 24.0 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 1.0 | 4.9 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.9 | 4.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.6 | 10.5 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.6 | 6.1 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.5 | 9.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.4 | 1.5 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.3 | 2.8 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.3 | 5.3 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.3 | 8.8 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.2 | 1.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 1.0 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.2 | 0.8 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.2 | 6.2 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.2 | 5.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 5.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.2 | 3.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 8.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.4 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.1 | 16.1 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.1 | 2.1 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 1.4 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 1.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 13.2 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 1.5 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 7.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.8 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.1 | 4.0 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 6.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 4.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 1.5 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 0.6 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 1.4 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 1.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 3.8 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 2.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.4 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 2.3 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.9 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 1.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 3.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 3.2 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 3.0 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 1.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 2.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 2.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 2.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.1 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.4 | 7.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.3 | 4.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 5.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 2.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.2 | 5.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.9 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 6.1 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.1 | 5.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 2.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 5.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.9 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 2.3 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.9 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 3.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |