Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
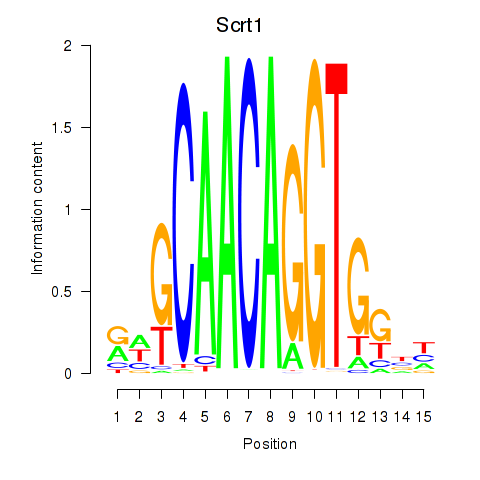
Results for Scrt1
Z-value: 1.14
Transcription factors associated with Scrt1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Scrt1
|
ENSMUSG00000048385.10 | Scrt1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Scrt1 | mm39_v1_chr15_-_76406102_76406258 | -0.80 | 4.2e-17 | Click! |
Activity profile of Scrt1 motif
Sorted Z-values of Scrt1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Scrt1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_83637766 | 8.68 |
ENSMUST00000070832.3
|
Wfdc21
|
WAP four-disulfide core domain 21 |
| chr1_-_192880260 | 8.11 |
ENSMUST00000161367.2
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr7_+_126690525 | 7.49 |
ENSMUST00000056288.7
ENSMUST00000206102.2 |
AI467606
|
expressed sequence AI467606 |
| chr11_-_83421333 | 7.22 |
ENSMUST00000035938.3
|
Ccl5
|
chemokine (C-C motif) ligand 5 |
| chr11_+_83007635 | 7.09 |
ENSMUST00000037994.8
|
Slfn1
|
schlafen 1 |
| chr10_+_79852487 | 6.43 |
ENSMUST00000099501.10
|
Arhgap45
|
Rho GTPase activating protein 45 |
| chr2_-_163592127 | 6.41 |
ENSMUST00000017841.4
|
Ada
|
adenosine deaminase |
| chr6_+_48695535 | 6.40 |
ENSMUST00000127537.6
ENSMUST00000052503.8 |
Gm28053
Gimap7
|
predicted gene, 28053 GTPase, IMAP family member 7 |
| chr11_+_53661251 | 6.31 |
ENSMUST00000138913.8
ENSMUST00000123376.8 ENSMUST00000019043.13 ENSMUST00000133291.3 |
Irf1
|
interferon regulatory factor 1 |
| chr19_+_11514132 | 5.90 |
ENSMUST00000025581.7
|
Ms4a4d
|
membrane-spanning 4-domains, subfamily A, member 4D |
| chr8_-_106863521 | 5.34 |
ENSMUST00000115979.9
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr15_+_77583104 | 5.25 |
ENSMUST00000096358.6
|
Apol7e
|
apolipoprotein L 7e |
| chr8_-_106863423 | 5.13 |
ENSMUST00000146940.2
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr19_+_24976864 | 5.11 |
ENSMUST00000025831.8
|
Dock8
|
dedicator of cytokinesis 8 |
| chr13_+_19528728 | 5.00 |
ENSMUST00000179181.3
|
Trgc4
|
T cell receptor gamma, constant 4 |
| chr9_-_31043076 | 4.97 |
ENSMUST00000034478.3
|
St14
|
suppression of tumorigenicity 14 (colon carcinoma) |
| chr6_+_78402956 | 4.92 |
ENSMUST00000079926.6
|
Reg1
|
regenerating islet-derived 1 |
| chr17_+_44499451 | 4.78 |
ENSMUST00000024755.7
|
Clic5
|
chloride intracellular channel 5 |
| chr9_+_110709301 | 4.70 |
ENSMUST00000123389.8
|
Als2cl
|
ALS2 C-terminal like |
| chr9_+_7764042 | 4.65 |
ENSMUST00000052865.16
|
Tmem123
|
transmembrane protein 123 |
| chr2_+_127178072 | 4.65 |
ENSMUST00000028846.7
|
Dusp2
|
dual specificity phosphatase 2 |
| chr19_+_37425180 | 4.56 |
ENSMUST00000128184.3
|
Hhex
|
hematopoietically expressed homeobox |
| chr1_+_85528392 | 4.43 |
ENSMUST00000080204.11
|
Sp140
|
Sp140 nuclear body protein |
| chr1_-_173770010 | 4.34 |
ENSMUST00000042228.15
ENSMUST00000081216.12 ENSMUST00000129829.8 ENSMUST00000123708.8 |
Ifi203
|
interferon activated gene 203 |
| chr15_+_79775894 | 4.34 |
ENSMUST00000177483.8
|
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr7_+_109820904 | 4.33 |
ENSMUST00000033325.9
|
Swap70
|
SWA-70 protein |
| chr16_-_4950285 | 4.32 |
ENSMUST00000035672.5
|
Ppl
|
periplakin |
| chr15_-_66432938 | 4.28 |
ENSMUST00000048372.7
|
Tmem71
|
transmembrane protein 71 |
| chr1_+_171594690 | 4.26 |
ENSMUST00000015460.5
|
Slamf1
|
signaling lymphocytic activation molecule family member 1 |
| chr2_+_152468850 | 4.16 |
ENSMUST00000000369.4
ENSMUST00000150913.2 |
Rem1
|
rad and gem related GTP binding protein 1 |
| chr6_+_138117519 | 4.11 |
ENSMUST00000120939.8
ENSMUST00000204628.3 ENSMUST00000140932.2 ENSMUST00000120302.8 |
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr7_-_30672824 | 3.97 |
ENSMUST00000147431.2
ENSMUST00000098553.11 ENSMUST00000108116.10 |
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr15_+_78209920 | 3.97 |
ENSMUST00000230264.3
|
Csf2rb
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr15_+_44320918 | 3.96 |
ENSMUST00000038336.12
ENSMUST00000166957.2 ENSMUST00000209244.2 |
Pkhd1l1
|
polycystic kidney and hepatic disease 1-like 1 |
| chr1_-_69723316 | 3.89 |
ENSMUST00000190855.7
ENSMUST00000188110.7 ENSMUST00000191262.7 |
Ikzf2
|
IKAROS family zinc finger 2 |
| chr15_+_78210190 | 3.88 |
ENSMUST00000229034.2
ENSMUST00000096355.4 |
Csf2rb
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr10_-_75768302 | 3.84 |
ENSMUST00000120281.8
ENSMUST00000000924.13 |
Mmp11
|
matrix metallopeptidase 11 |
| chr10_-_81127334 | 3.83 |
ENSMUST00000219479.2
|
Tjp3
|
tight junction protein 3 |
| chr4_+_102976472 | 3.77 |
ENSMUST00000106857.8
|
Mier1
|
MEIR1 treanscription regulator |
| chr6_+_138118565 | 3.73 |
ENSMUST00000118091.8
|
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr9_-_14815228 | 3.71 |
ENSMUST00000034409.14
ENSMUST00000117620.8 |
Izumo1r
|
IZUMO1 receptor, JUNO |
| chrX_-_71318353 | 3.68 |
ENSMUST00000064780.4
|
Gabre
|
gamma-aminobutyric acid (GABA) A receptor, subunit epsilon |
| chr2_+_153334710 | 3.60 |
ENSMUST00000109783.2
|
4930404H24Rik
|
RIKEN cDNA 4930404H24 gene |
| chr6_+_41025217 | 3.58 |
ENSMUST00000103264.3
|
Trbv3
|
T cell receptor beta, variable 3 |
| chr7_-_30672889 | 3.57 |
ENSMUST00000001279.15
|
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr2_+_131075965 | 3.56 |
ENSMUST00000041362.12
ENSMUST00000110199.3 |
Mavs
|
mitochondrial antiviral signaling protein |
| chr8_-_71990085 | 3.55 |
ENSMUST00000051672.9
|
Bst2
|
bone marrow stromal cell antigen 2 |
| chr10_-_81127057 | 3.52 |
ENSMUST00000045744.7
|
Tjp3
|
tight junction protein 3 |
| chr17_-_56019571 | 3.50 |
ENSMUST00000086876.7
ENSMUST00000233557.2 ENSMUST00000233972.2 |
Pot1b
|
protection of telomeres 1B |
| chr18_+_11790409 | 3.50 |
ENSMUST00000047322.8
|
Rbbp8
|
retinoblastoma binding protein 8, endonuclease |
| chr1_-_173707677 | 3.48 |
ENSMUST00000190651.4
ENSMUST00000188804.7 |
Mndal
|
myeloid nuclear differentiation antigen like |
| chr6_+_48695571 | 3.46 |
ENSMUST00000204785.2
|
Gimap7
|
GTPase, IMAP family member 7 |
| chr15_+_78210242 | 3.42 |
ENSMUST00000229678.2
ENSMUST00000231888.2 |
Csf2rb
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr11_-_101357046 | 3.37 |
ENSMUST00000040430.8
|
Vat1
|
vesicle amine transport 1 |
| chr15_-_77331660 | 3.36 |
ENSMUST00000089469.7
|
Apol7b
|
apolipoprotein L 7b |
| chr9_+_110709260 | 3.36 |
ENSMUST00000130386.8
|
Als2cl
|
ALS2 C-terminal like |
| chr4_-_11386394 | 3.34 |
ENSMUST00000155519.2
|
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr7_+_140547941 | 3.32 |
ENSMUST00000106040.8
ENSMUST00000026564.9 |
Ifitm1
|
interferon induced transmembrane protein 1 |
| chrX_-_132882514 | 3.28 |
ENSMUST00000113297.9
ENSMUST00000174542.2 ENSMUST00000033608.15 ENSMUST00000113294.8 |
Sytl4
|
synaptotagmin-like 4 |
| chr8_+_72943455 | 3.24 |
ENSMUST00000072097.14
|
Hsh2d
|
hematopoietic SH2 domain containing |
| chr5_+_33815466 | 3.24 |
ENSMUST00000074849.13
ENSMUST00000079534.11 ENSMUST00000201633.2 |
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr5_+_137627431 | 3.23 |
ENSMUST00000176667.8
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr4_-_57916283 | 3.20 |
ENSMUST00000063816.6
|
D630039A03Rik
|
RIKEN cDNA D630039A03 gene |
| chr1_+_131794962 | 3.14 |
ENSMUST00000112386.8
ENSMUST00000027693.8 |
Rab29
|
RAB29, member RAS oncogene family |
| chr9_+_110709223 | 3.10 |
ENSMUST00000084926.9
|
Als2cl
|
ALS2 C-terminal like |
| chr8_-_123338157 | 3.04 |
ENSMUST00000015171.11
|
Galns
|
galactosamine (N-acetyl)-6-sulfate sulfatase |
| chr6_-_41012435 | 3.01 |
ENSMUST00000031931.6
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr9_+_110709353 | 3.00 |
ENSMUST00000155014.2
|
Als2cl
|
ALS2 C-terminal like |
| chr6_-_88875646 | 2.97 |
ENSMUST00000058011.8
|
Mcm2
|
minichromosome maintenance complex component 2 |
| chr12_-_115083839 | 2.96 |
ENSMUST00000103521.3
|
Ighv1-50
|
immunoglobulin heavy variable 1-50 |
| chr10_-_61814852 | 2.93 |
ENSMUST00000105453.8
ENSMUST00000105452.9 ENSMUST00000105454.3 |
Col13a1
|
collagen, type XIII, alpha 1 |
| chr7_+_13011180 | 2.92 |
ENSMUST00000177588.10
|
Lig1
|
ligase I, DNA, ATP-dependent |
| chr14_+_53994813 | 2.88 |
ENSMUST00000180380.3
|
Trav13-4-dv7
|
T cell receptor alpha variable 13-4-DV7 |
| chr4_-_129083439 | 2.88 |
ENSMUST00000106059.8
|
S100pbp
|
S100P binding protein |
| chr2_+_150751475 | 2.88 |
ENSMUST00000028948.5
|
Gins1
|
GINS complex subunit 1 (Psf1 homolog) |
| chr11_+_68447012 | 2.87 |
ENSMUST00000053211.8
|
Mfsd6l
|
major facilitator superfamily domain containing 6-like |
| chr5_+_137628377 | 2.87 |
ENSMUST00000175968.8
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr2_-_104680104 | 2.87 |
ENSMUST00000028593.11
|
Prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr4_-_129083335 | 2.86 |
ENSMUST00000106061.9
ENSMUST00000072431.13 |
S100pbp
|
S100P binding protein |
| chr17_+_34879431 | 2.83 |
ENSMUST00000238967.2
|
Tnxb
|
tenascin XB |
| chr12_-_115876396 | 2.76 |
ENSMUST00000103547.2
|
Ighv1-80
|
immunoglobulin heavy variable 1-80 |
| chr5_-_137145030 | 2.76 |
ENSMUST00000054384.6
ENSMUST00000152207.2 |
Trim56
|
tripartite motif-containing 56 |
| chr9_+_92132345 | 2.75 |
ENSMUST00000186364.2
|
Plscr1
|
phospholipid scramblase 1 |
| chr19_-_6835538 | 2.74 |
ENSMUST00000113440.2
|
Ccdc88b
|
coiled-coil domain containing 88B |
| chr1_-_85248264 | 2.74 |
ENSMUST00000093506.12
ENSMUST00000064341.9 |
C130026I21Rik
|
RIKEN cDNA C130026I21 gene |
| chr4_-_129083251 | 2.72 |
ENSMUST00000117965.8
|
S100pbp
|
S100P binding protein |
| chr13_+_22129246 | 2.72 |
ENSMUST00000176511.8
ENSMUST00000102978.8 ENSMUST00000152258.9 |
Zfp184
|
zinc finger protein 184 (Kruppel-like) |
| chr12_-_115884332 | 2.72 |
ENSMUST00000103548.3
|
Ighv1-81
|
immunoglobulin heavy variable 1-81 |
| chr19_+_6414587 | 2.72 |
ENSMUST00000155973.2
|
Sf1
|
splicing factor 1 |
| chr8_-_123338126 | 2.71 |
ENSMUST00000212319.2
|
Galns
|
galactosamine (N-acetyl)-6-sulfate sulfatase |
| chr1_-_156767196 | 2.69 |
ENSMUST00000185198.7
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr17_-_15181491 | 2.67 |
ENSMUST00000024657.12
|
Phf10
|
PHD finger protein 10 |
| chr4_-_129083392 | 2.66 |
ENSMUST00000117497.8
ENSMUST00000117350.2 |
S100pbp
|
S100P binding protein |
| chr7_+_140343652 | 2.64 |
ENSMUST00000026552.9
ENSMUST00000209253.2 ENSMUST00000210235.2 |
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr7_-_30672747 | 2.64 |
ENSMUST00000205961.2
|
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr4_-_115980813 | 2.62 |
ENSMUST00000102704.4
ENSMUST00000102705.10 |
Rad54l
|
RAD54 like (S. cerevisiae) |
| chr7_+_79944198 | 2.58 |
ENSMUST00000163812.9
ENSMUST00000047558.14 ENSMUST00000174199.8 ENSMUST00000173824.8 ENSMUST00000174172.8 |
Prc1
|
protein regulator of cytokinesis 1 |
| chr6_-_120334382 | 2.58 |
ENSMUST00000032283.12
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr15_-_96947963 | 2.57 |
ENSMUST00000230907.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr5_-_138170077 | 2.56 |
ENSMUST00000155902.8
ENSMUST00000148879.8 |
Mcm7
|
minichromosome maintenance complex component 7 |
| chr15_+_79775819 | 2.55 |
ENSMUST00000177350.8
|
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr7_+_30673212 | 2.54 |
ENSMUST00000129773.2
|
Fam187b
|
family with sequence similarity 187, member B |
| chr10_-_127843377 | 2.52 |
ENSMUST00000219447.2
ENSMUST00000219780.2 ENSMUST00000219707.2 ENSMUST00000219953.2 ENSMUST00000219183.2 |
Hsd17b6
|
hydroxysteroid (17-beta) dehydrogenase 6 |
| chr17_+_24257118 | 2.50 |
ENSMUST00000059482.6
|
Prss27
|
protease, serine 27 |
| chr4_-_19570073 | 2.49 |
ENSMUST00000029885.5
|
Cpne3
|
copine III |
| chr3_-_123029782 | 2.49 |
ENSMUST00000106427.8
ENSMUST00000198584.2 |
Synpo2
|
synaptopodin 2 |
| chr15_-_100576715 | 2.48 |
ENSMUST00000229869.2
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr15_+_74828272 | 2.47 |
ENSMUST00000188042.2
|
Ly6e
|
lymphocyte antigen 6 complex, locus E |
| chr18_+_24338729 | 2.47 |
ENSMUST00000170243.8
|
Galnt1
|
polypeptide N-acetylgalactosaminyltransferase 1 |
| chr8_+_72889607 | 2.46 |
ENSMUST00000238492.2
|
Tpm4
|
tropomyosin 4 |
| chr15_-_100583044 | 2.46 |
ENSMUST00000230312.2
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr6_+_21985902 | 2.46 |
ENSMUST00000115383.9
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr5_-_137609634 | 2.43 |
ENSMUST00000054564.13
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr5_+_33815910 | 2.39 |
ENSMUST00000114426.10
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr3_-_123029745 | 2.38 |
ENSMUST00000106426.8
|
Synpo2
|
synaptopodin 2 |
| chr2_-_104679838 | 2.37 |
ENSMUST00000126824.2
|
Prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr3_-_132797558 | 2.37 |
ENSMUST00000029651.11
|
Gstcd
|
glutathione S-transferase, C-terminal domain containing |
| chr5_-_137609691 | 2.35 |
ENSMUST00000031731.14
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr5_+_38434738 | 2.35 |
ENSMUST00000114099.6
|
Otop1
|
otopetrin 1 |
| chr10_+_78412783 | 2.34 |
ENSMUST00000219588.2
|
Ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr10_-_111829393 | 2.32 |
ENSMUST00000161870.3
|
Glipr1
|
GLI pathogenesis-related 1 (glioma) |
| chr6_-_120334302 | 2.32 |
ENSMUST00000163827.8
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr12_-_115811951 | 2.31 |
ENSMUST00000197537.2
|
Ighv1-76
|
immunoglobulin heavy variable 1-76 |
| chr6_-_120334400 | 2.31 |
ENSMUST00000112703.8
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr3_-_108747792 | 2.30 |
ENSMUST00000106596.4
ENSMUST00000102621.11 ENSMUST00000138552.6 |
Stxbp3
|
syntaxin binding protein 3 |
| chr7_+_24990596 | 2.29 |
ENSMUST00000164820.2
|
Cic
|
capicua transcriptional repressor |
| chr7_-_105249308 | 2.27 |
ENSMUST00000210531.2
ENSMUST00000033185.10 |
Hpx
|
hemopexin |
| chr5_-_144294854 | 2.26 |
ENSMUST00000055190.8
|
Baiap2l1
|
BAI1-associated protein 2-like 1 |
| chr19_+_29229826 | 2.26 |
ENSMUST00000238009.2
|
Jak2
|
Janus kinase 2 |
| chr6_+_122967309 | 2.25 |
ENSMUST00000079379.3
|
Clec4a4
|
C-type lectin domain family 4, member a4 |
| chr2_-_60711706 | 2.22 |
ENSMUST00000164147.8
ENSMUST00000112509.2 |
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr6_-_86646118 | 2.22 |
ENSMUST00000001184.10
|
Mxd1
|
MAX dimerization protein 1 |
| chr18_-_14815807 | 2.18 |
ENSMUST00000040964.13
ENSMUST00000234524.2 ENSMUST00000092041.11 ENSMUST00000040924.9 |
Ss18
|
SS18, nBAF chromatin remodeling complex subunit |
| chr6_+_127538268 | 2.17 |
ENSMUST00000212051.2
|
Cracr2a
|
calcium release activated channel regulator 2A |
| chr7_-_81642034 | 2.16 |
ENSMUST00000026096.10
|
Bnc1
|
basonuclin 1 |
| chr8_+_47192911 | 2.14 |
ENSMUST00000208433.2
|
Irf2
|
interferon regulatory factor 2 |
| chr1_-_156767123 | 2.11 |
ENSMUST00000189316.7
ENSMUST00000190648.7 ENSMUST00000172057.8 ENSMUST00000191605.7 |
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr19_-_42741148 | 2.08 |
ENSMUST00000236659.2
ENSMUST00000076505.4 |
Pyroxd2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr7_-_28981335 | 2.08 |
ENSMUST00000108236.5
ENSMUST00000098604.12 |
Spint2
|
serine protease inhibitor, Kunitz type 2 |
| chr5_-_138169509 | 2.04 |
ENSMUST00000153867.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr8_+_4399588 | 2.01 |
ENSMUST00000110982.8
ENSMUST00000024004.9 |
Ccl25
|
chemokine (C-C motif) ligand 25 |
| chr2_+_72115981 | 2.00 |
ENSMUST00000090824.12
ENSMUST00000135469.8 |
Map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr7_+_27559990 | 1.99 |
ENSMUST00000205534.2
|
Zfp607a
|
zinc finger protein 607A |
| chr6_-_116650751 | 1.98 |
ENSMUST00000204576.2
ENSMUST00000203029.3 ENSMUST00000035842.7 |
Rassf4
|
Ras association (RalGDS/AF-6) domain family member 4 |
| chr6_+_78382131 | 1.98 |
ENSMUST00000023906.4
|
Reg2
|
regenerating islet-derived 2 |
| chr3_+_123061094 | 1.96 |
ENSMUST00000047923.12
ENSMUST00000200333.2 |
Sec24d
|
Sec24 related gene family, member D (S. cerevisiae) |
| chr1_-_156766957 | 1.95 |
ENSMUST00000171292.8
ENSMUST00000063199.13 ENSMUST00000027886.14 |
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr18_+_60880149 | 1.93 |
ENSMUST00000236652.2
ENSMUST00000235966.2 |
Rps14
|
ribosomal protein S14 |
| chr3_-_108747767 | 1.90 |
ENSMUST00000196679.5
|
Stxbp3
|
syntaxin binding protein 3 |
| chr15_-_5137975 | 1.88 |
ENSMUST00000118365.3
|
Card6
|
caspase recruitment domain family, member 6 |
| chr12_+_65083093 | 1.88 |
ENSMUST00000120580.8
|
Prpf39
|
pre-mRNA processing factor 39 |
| chr14_+_31750946 | 1.88 |
ENSMUST00000022460.11
|
Galnt15
|
polypeptide N-acetylgalactosaminyltransferase 15 |
| chr3_+_10431961 | 1.86 |
ENSMUST00000029049.7
|
Chmp4c
|
charged multivesicular body protein 4C |
| chr10_-_106959462 | 1.85 |
ENSMUST00000044668.5
|
Acss3
|
acyl-CoA synthetase short-chain family member 3 |
| chr14_+_54412058 | 1.83 |
ENSMUST00000103699.2
|
Traj43
|
T cell receptor alpha joining 43 |
| chr16_+_36514334 | 1.83 |
ENSMUST00000023617.13
ENSMUST00000089618.10 |
Ildr1
|
immunoglobulin-like domain containing receptor 1 |
| chr9_+_65122381 | 1.82 |
ENSMUST00000213396.2
|
Parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr7_-_100504610 | 1.82 |
ENSMUST00000156855.8
|
Relt
|
RELT tumor necrosis factor receptor |
| chr16_+_31998588 | 1.79 |
ENSMUST00000231941.2
|
Bex6
|
brain expressed family member 6 |
| chr8_+_91635192 | 1.78 |
ENSMUST00000211403.2
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr18_+_24338358 | 1.77 |
ENSMUST00000000430.14
|
Galnt1
|
polypeptide N-acetylgalactosaminyltransferase 1 |
| chr17_-_35334404 | 1.77 |
ENSMUST00000172854.2
ENSMUST00000062657.5 |
Ly6g5b
|
lymphocyte antigen 6 complex, locus G5B |
| chr3_+_132797588 | 1.76 |
ENSMUST00000029650.9
|
Ints12
|
integrator complex subunit 12 |
| chr1_+_172309337 | 1.76 |
ENSMUST00000127052.8
|
Igsf9
|
immunoglobulin superfamily, member 9 |
| chr9_-_14815163 | 1.75 |
ENSMUST00000069408.10
|
Izumo1r
|
IZUMO1 receptor, JUNO |
| chr12_-_114576295 | 1.74 |
ENSMUST00000191801.2
|
Ighv1-11
|
immunoglobulin heavy variable V1-11 |
| chr9_+_78164402 | 1.72 |
ENSMUST00000217203.2
|
Gm3776
|
predicted gene 3776 |
| chr3_-_132940647 | 1.71 |
ENSMUST00000147041.10
ENSMUST00000161022.9 |
Arhgef38
|
Rho guanine nucleotide exchange factor (GEF) 38 |
| chr6_+_123149847 | 1.70 |
ENSMUST00000088455.6
|
Clec4b2
|
C-type lectin domain family 4, member b2 |
| chr13_-_34837469 | 1.67 |
ENSMUST00000053459.15
|
Pxdc1
|
PX domain containing 1 |
| chr10_-_106959444 | 1.65 |
ENSMUST00000165067.9
|
Acss3
|
acyl-CoA synthetase short-chain family member 3 |
| chr2_-_29983618 | 1.59 |
ENSMUST00000081838.7
ENSMUST00000102865.11 |
Zdhhc12
|
zinc finger, DHHC domain containing 12 |
| chr1_-_156131155 | 1.59 |
ENSMUST00000141760.4
ENSMUST00000121146.10 |
Tdrd5
|
tudor domain containing 5 |
| chr5_+_33815892 | 1.57 |
ENSMUST00000152847.8
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chrX_+_55181906 | 1.56 |
ENSMUST00000033458.3
|
3830403N18Rik
|
RIKEN cDNA 3830403N18 gene |
| chr12_+_113061819 | 1.51 |
ENSMUST00000109727.9
ENSMUST00000009099.13 ENSMUST00000109723.8 ENSMUST00000109726.8 ENSMUST00000069690.5 |
Mta1
|
metastasis associated 1 |
| chr7_-_30623592 | 1.51 |
ENSMUST00000217812.2
ENSMUST00000074671.9 |
Hamp2
|
hepcidin antimicrobial peptide 2 |
| chr6_+_88061464 | 1.49 |
ENSMUST00000032143.8
|
Rpn1
|
ribophorin I |
| chr4_-_93223746 | 1.49 |
ENSMUST00000066774.6
|
Tusc1
|
tumor suppressor candidate 1 |
| chr2_-_169984593 | 1.48 |
ENSMUST00000109155.8
|
Zfp217
|
zinc finger protein 217 |
| chr5_+_146769700 | 1.48 |
ENSMUST00000035983.12
|
Rpl21
|
ribosomal protein L21 |
| chr5_+_90933953 | 1.47 |
ENSMUST00000031326.10
|
Cxcl3
|
chemokine (C-X-C motif) ligand 3 |
| chr12_+_87193922 | 1.47 |
ENSMUST00000222885.2
|
Gstz1
|
glutathione transferase zeta 1 (maleylacetoacetate isomerase) |
| chr12_-_86931529 | 1.47 |
ENSMUST00000038422.8
|
Irf2bpl
|
interferon regulatory factor 2 binding protein-like |
| chr11_+_70396070 | 1.47 |
ENSMUST00000019063.3
|
Tm4sf5
|
transmembrane 4 superfamily member 5 |
| chr3_-_94693780 | 1.46 |
ENSMUST00000107273.9
ENSMUST00000238849.2 |
Cgn
|
cingulin |
| chr9_+_65121944 | 1.46 |
ENSMUST00000069000.9
|
Parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr11_-_114851243 | 1.46 |
ENSMUST00000092466.13
ENSMUST00000061637.4 |
Cd300c
|
CD300C molecule |
| chr16_+_36514386 | 1.44 |
ENSMUST00000119464.2
|
Ildr1
|
immunoglobulin-like domain containing receptor 1 |
| chr7_+_141027557 | 1.44 |
ENSMUST00000106004.8
|
Rplp2
|
ribosomal protein, large P2 |
| chr1_+_88154727 | 1.43 |
ENSMUST00000061013.13
ENSMUST00000113130.8 |
Mroh2a
|
maestro heat-like repeat family member 2A |
| chr2_-_164753480 | 1.43 |
ENSMUST00000041361.14
|
Zfp335
|
zinc finger protein 335 |
| chr6_-_116650728 | 1.42 |
ENSMUST00000204203.2
|
Rassf4
|
Ras association (RalGDS/AF-6) domain family member 4 |
| chr15_-_78189917 | 1.42 |
ENSMUST00000096356.5
|
Csf2rb2
|
colony stimulating factor 2 receptor, beta 2, low-affinity (granulocyte-macrophage) |
| chr11_+_115225557 | 1.41 |
ENSMUST00000106543.8
ENSMUST00000019006.5 |
Otop3
|
otopetrin 3 |
| chr9_-_110709175 | 1.39 |
ENSMUST00000050958.9
|
Tmie
|
transmembrane inner ear |
| chr17_+_34573760 | 1.39 |
ENSMUST00000178562.2
ENSMUST00000025198.15 |
Btnl2
|
butyrophilin-like 2 |
| chr3_-_94693740 | 1.38 |
ENSMUST00000153263.9
ENSMUST00000107272.7 ENSMUST00000155485.4 |
Cgn
|
cingulin |
| chr11_-_6394352 | 1.38 |
ENSMUST00000093346.6
|
H2az2
|
H2A.Z histone variant 2 |
| chr16_+_48637219 | 1.38 |
ENSMUST00000023328.8
|
Retnlb
|
resistin like beta |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 10.2 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 2.4 | 7.1 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 2.3 | 7.0 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 2.1 | 6.4 | GO:0046100 | germinal center B cell differentiation(GO:0002314) adenosine catabolic process(GO:0006154) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) purine deoxyribonucleoside metabolic process(GO:0046122) negative regulation of mucus secretion(GO:0070256) |
| 2.0 | 7.8 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 1.8 | 7.2 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 1.6 | 6.3 | GO:0035740 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 1.4 | 4.3 | GO:0035744 | myeloid dendritic cell activation involved in immune response(GO:0002277) T-helper 1 cell cytokine production(GO:0035744) |
| 1.2 | 3.6 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 1.2 | 3.5 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 1.1 | 4.3 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.8 | 3.4 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.8 | 2.3 | GO:0009629 | response to gravity(GO:0009629) |
| 0.8 | 2.3 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.7 | 3.6 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.7 | 4.2 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.7 | 4.9 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.7 | 4.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.7 | 2.8 | GO:2000371 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.7 | 6.7 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.7 | 4.0 | GO:0061017 | hepatoblast differentiation(GO:0061017) |
| 0.7 | 8.0 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.6 | 1.9 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.5 | 3.3 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.5 | 3.2 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.5 | 1.6 | GO:0007314 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 0.5 | 5.8 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.5 | 2.0 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.5 | 8.1 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.4 | 2.6 | GO:1903436 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.4 | 2.5 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.4 | 3.5 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.4 | 1.5 | GO:0034757 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 0.4 | 0.7 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.4 | 8.6 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.3 | 2.4 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.3 | 6.9 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.3 | 2.3 | GO:0060335 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) heme transport(GO:0015886) positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.3 | 4.3 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.3 | 1.2 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.3 | 13.8 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.3 | 1.8 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.3 | 1.5 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.3 | 8.7 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.3 | 5.5 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.3 | 2.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.3 | 4.2 | GO:0022615 | protein to membrane docking(GO:0022615) |
| 0.3 | 14.2 | GO:0007032 | endosome organization(GO:0007032) |
| 0.3 | 4.6 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.3 | 2.2 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.3 | 3.8 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.3 | 6.1 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.3 | 1.3 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.2 | 0.7 | GO:0021762 | substantia nigra development(GO:0021762) corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.2 | 3.8 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.2 | 0.9 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.2 | 1.1 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.2 | 0.4 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.2 | 0.9 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.2 | 0.7 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.2 | 3.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 1.3 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.2 | 1.1 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.2 | 3.1 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.2 | 1.9 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 0.4 | GO:0060775 | mediolateral intercalation(GO:0060031) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) |
| 0.2 | 5.1 | GO:0036336 | dendritic cell migration(GO:0036336) |
| 0.2 | 0.6 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.2 | 1.2 | GO:0071442 | N-terminal peptidyl-lysine acetylation(GO:0018076) positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.2 | 1.7 | GO:0002579 | positive regulation of antigen processing and presentation(GO:0002579) |
| 0.2 | 4.6 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.2 | 3.9 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.2 | 2.5 | GO:0048242 | epinephrine secretion(GO:0048242) |
| 0.2 | 9.0 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.2 | 1.5 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 0.6 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.2 | 0.5 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.1 | 3.8 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 1.2 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 0.7 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 3.5 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.1 | 1.2 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.4 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.5 | GO:2000660 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 4.0 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 1.1 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.1 | 0.3 | GO:0046333 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.1 | 0.4 | GO:0072021 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.1 | 4.3 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 0.6 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.1 | 0.3 | GO:0021508 | floor plate formation(GO:0021508) |
| 0.1 | 0.7 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 1.5 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 1.4 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 1.9 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 4.6 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.1 | 0.7 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 1.4 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 2.9 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 2.0 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.1 | 3.7 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 12.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 4.3 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.1 | 2.7 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.1 | 2.3 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.4 | GO:0052428 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.1 | 1.1 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 1.3 | GO:0097012 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.1 | 0.3 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 1.9 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 1.0 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 0.3 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.1 | 1.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.5 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.1 | 1.2 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.2 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 1.3 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.8 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.4 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 2.2 | GO:0006356 | regulation of transcription from RNA polymerase I promoter(GO:0006356) |
| 0.0 | 2.0 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.1 | GO:1901662 | menaquinone metabolic process(GO:0009233) phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 2.6 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 1.0 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 8.0 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 6.3 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 5.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.5 | GO:0003093 | regulation of glomerular filtration(GO:0003093) |
| 0.0 | 0.8 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 1.2 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 1.5 | GO:0031050 | dsRNA fragmentation(GO:0031050) production of miRNAs involved in gene silencing by miRNA(GO:0035196) production of small RNA involved in gene silencing by RNA(GO:0070918) |
| 0.0 | 0.5 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.8 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.8 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.6 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.2 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 1.0 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 2.3 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.4 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.2 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 | 0.4 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 1.5 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.6 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 8.7 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 1.4 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.3 | GO:0032056 | positive regulation of translation in response to stress(GO:0032056) |
| 0.0 | 1.4 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.7 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.0 | 0.8 | GO:0042990 | regulation of transcription factor import into nucleus(GO:0042990) |
| 0.0 | 3.9 | GO:0043122 | regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043122) |
| 0.0 | 5.0 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.1 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 1.1 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.0 | 0.3 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.0 | 0.5 | GO:0042168 | heme metabolic process(GO:0042168) |
| 0.0 | 1.0 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.2 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.0 | 0.5 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 13.4 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 1.0 | 2.9 | GO:0000811 | GINS complex(GO:0000811) |
| 0.7 | 2.0 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.6 | 4.2 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.6 | 9.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.3 | 0.9 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.2 | 3.5 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.2 | 4.3 | GO:0090543 | Flemming body(GO:0090543) |
| 0.2 | 1.3 | GO:0032807 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.2 | 3.1 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 0.2 | 4.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 2.5 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 1.4 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.2 | 2.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.2 | 2.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.6 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 6.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.4 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 1.9 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 1.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 11.4 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 4.1 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 12.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 3.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 3.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 2.6 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 0.6 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 0.4 | GO:0002142 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.1 | 0.7 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 8.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.5 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 10.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 4.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 2.9 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 4.6 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 1.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 1.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.4 | GO:0044299 | C-fiber(GO:0044299) |
| 0.1 | 1.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.8 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.5 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 3.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 6.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 2.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 6.8 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.8 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 4.3 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.6 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 1.0 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 4.6 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.2 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 4.6 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.7 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 3.4 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 11.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 4.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 4.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 1.4 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 7.0 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 2.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 8.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.0 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 13.4 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 2.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 3.6 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 0.4 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.8 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 20.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 2.0 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 0.2 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 1.1 | 6.9 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 1.0 | 3.9 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.7 | 3.5 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.7 | 2.7 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.5 | 2.0 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.5 | 1.5 | GO:0016034 | maleylacetoacetate isomerase activity(GO:0016034) |
| 0.5 | 1.9 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.5 | 6.4 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.5 | 2.3 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.4 | 1.3 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.4 | 3.5 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.4 | 10.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.4 | 1.3 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.4 | 4.2 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.4 | 2.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.3 | 1.3 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.3 | 1.0 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.3 | 5.7 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.3 | 2.5 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.3 | 3.6 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.3 | 4.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.3 | 6.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.3 | 4.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.3 | 7.8 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.3 | 1.8 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.3 | 14.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 3.5 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.2 | 2.5 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 11.8 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.2 | 2.6 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.2 | 0.9 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.2 | 1.5 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.2 | 2.8 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.2 | 2.3 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.2 | 1.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 1.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.2 | 5.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 5.5 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.2 | 3.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 1.9 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.2 | 1.1 | GO:0031013 | troponin C binding(GO:0030172) calcium-dependent ATPase activity(GO:0030899) troponin I binding(GO:0031013) |
| 0.1 | 1.5 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 3.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.6 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 1.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 1.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 5.2 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 0.4 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.1 | 3.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 13.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 4.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.5 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 3.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 1.9 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.1 | 0.3 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.1 | 1.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.8 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.7 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 1.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 12.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.6 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 3.1 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 1.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 4.0 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 3.8 | GO:0033558 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.1 | 2.6 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 0.5 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 3.7 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 0.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 8.8 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 14.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.3 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.1 | 2.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.8 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 1.8 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 1.0 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 4.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 1.0 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.1 | 3.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 4.8 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 2.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 8.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 14.7 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.4 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 1.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 4.9 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.9 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.8 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.4 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 1.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 2.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 8.3 | GO:0030414 | peptidase inhibitor activity(GO:0030414) peptidase regulator activity(GO:0061134) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 7.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.6 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 8.7 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 2.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 3.1 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 4.9 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 2.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 3.0 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.1 | GO:0047874 | dolichyldiphosphatase activity(GO:0047874) |
| 0.0 | 3.4 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.0 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 2.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 5.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 4.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.4 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.0 | 0.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.7 | GO:0019902 | phosphatase binding(GO:0019902) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 13.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.2 | 8.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 7.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 6.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 3.5 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 8.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 6.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 1.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 14.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.8 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 8.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.6 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.4 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 2.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 2.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.6 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.7 | 10.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.4 | 12.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.3 | 14.8 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.2 | 5.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 4.2 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.2 | 10.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 7.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 2.9 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 3.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 2.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 7.4 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.1 | 3.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 5.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 2.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 0.8 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.1 | 0.8 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 1.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 2.6 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.1 | 0.8 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 3.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 2.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 7.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 3.8 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.8 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 6.3 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.9 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 4.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.8 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.8 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 2.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |