Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Scrt2
Z-value: 1.37
Transcription factors associated with Scrt2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Scrt2
|
ENSMUSG00000060257.3 | Scrt2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Scrt2 | mm39_v1_chr2_+_151923449_151923544 | 0.78 | 8.4e-16 | Click! |
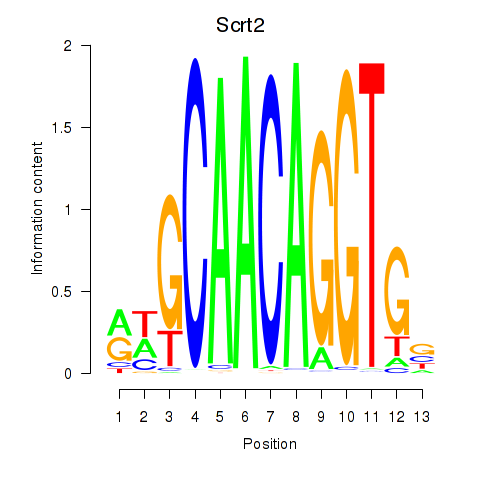
Activity profile of Scrt2 motif
Sorted Z-values of Scrt2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Scrt2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_66833434 | 19.73 |
ENSMUST00000031131.11
|
Uchl1
|
ubiquitin carboxy-terminal hydrolase L1 |
| chr9_+_45281483 | 18.72 |
ENSMUST00000085939.8
ENSMUST00000217381.2 |
Fxyd6
|
FXYD domain-containing ion transport regulator 6 |
| chr1_-_79417732 | 15.17 |
ENSMUST00000185234.2
ENSMUST00000049972.6 |
Scg2
|
secretogranin II |
| chr14_+_9646630 | 13.55 |
ENSMUST00000112658.8
ENSMUST00000112657.9 ENSMUST00000177814.2 ENSMUST00000067491.14 |
Cadps
|
Ca2+-dependent secretion activator |
| chr2_-_148574353 | 13.16 |
ENSMUST00000028926.13
|
Napb
|
N-ethylmaleimide sensitive fusion protein attachment protein beta |
| chrX_+_151922936 | 12.38 |
ENSMUST00000039720.11
ENSMUST00000144175.3 |
Rragb
|
Ras-related GTP binding B |
| chr11_-_100246209 | 12.24 |
ENSMUST00000146878.3
|
Hap1
|
huntingtin-associated protein 1 |
| chr5_+_37403098 | 12.22 |
ENSMUST00000031004.11
|
Crmp1
|
collapsin response mediator protein 1 |
| chr9_+_58395850 | 11.18 |
ENSMUST00000085658.5
|
Insyn1
|
inhibitory synaptic factor 1 |
| chr10_+_56253418 | 10.86 |
ENSMUST00000068581.9
ENSMUST00000217789.2 |
Gja1
|
gap junction protein, alpha 1 |
| chr13_+_97377604 | 10.65 |
ENSMUST00000041623.9
|
Enc1
|
ectodermal-neural cortex 1 |
| chr7_-_118047394 | 10.31 |
ENSMUST00000203796.4
ENSMUST00000203485.3 |
Syt17
|
synaptotagmin XVII |
| chr8_-_4309257 | 10.12 |
ENSMUST00000053252.9
|
Ctxn1
|
cortexin 1 |
| chr11_+_103540391 | 10.10 |
ENSMUST00000057870.4
|
Rprml
|
reprimo-like |
| chr5_+_118307754 | 9.84 |
ENSMUST00000054836.7
|
Hrk
|
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chr3_-_33197157 | 9.56 |
ENSMUST00000108226.8
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr10_-_79973210 | 9.18 |
ENSMUST00000170219.9
ENSMUST00000169546.9 |
Cbarp
|
calcium channel, voltage-dependent, beta subunit associated regulatory protein |
| chr3_+_129974531 | 9.12 |
ENSMUST00000080335.11
ENSMUST00000106353.2 |
Col25a1
|
collagen, type XXV, alpha 1 |
| chr2_-_53975501 | 9.02 |
ENSMUST00000100089.3
|
Rprm
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr2_-_73605387 | 8.80 |
ENSMUST00000166199.9
|
Chn1
|
chimerin 1 |
| chr11_+_71640739 | 8.10 |
ENSMUST00000150531.2
|
Wscd1
|
WSC domain containing 1 |
| chrX_+_5959507 | 7.81 |
ENSMUST00000103007.4
|
Nudt11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr16_+_16888145 | 7.39 |
ENSMUST00000232574.2
|
Ypel1
|
yippee like 1 |
| chr11_+_77384234 | 7.06 |
ENSMUST00000037285.10
ENSMUST00000100812.4 |
Git1
|
GIT ArfGAP 1 |
| chrX_-_100128958 | 6.79 |
ENSMUST00000101362.8
ENSMUST00000073927.5 |
Slc7a3
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3 |
| chr16_+_20514925 | 6.73 |
ENSMUST00000128273.2
|
Fam131a
|
family with sequence similarity 131, member A |
| chr14_+_55120777 | 6.26 |
ENSMUST00000022806.10
|
Bcl2l2
|
BCL2-like 2 |
| chr8_+_31677345 | 6.22 |
ENSMUST00000066173.12
ENSMUST00000161788.8 ENSMUST00000110527.9 ENSMUST00000161502.2 |
Fut10
|
fucosyltransferase 10 |
| chr10_-_79449826 | 5.98 |
ENSMUST00000059699.9
ENSMUST00000178228.3 |
C2cd4c
|
C2 calcium-dependent domain containing 4C |
| chr2_+_121188195 | 5.97 |
ENSMUST00000125812.8
ENSMUST00000078222.9 ENSMUST00000125221.3 ENSMUST00000150271.8 |
Ckmt1
|
creatine kinase, mitochondrial 1, ubiquitous |
| chr6_-_23839136 | 5.94 |
ENSMUST00000166458.9
ENSMUST00000142913.9 ENSMUST00000069074.14 ENSMUST00000115361.9 ENSMUST00000018122.14 ENSMUST00000115356.3 |
Cadps2
|
Ca2+-dependent activator protein for secretion 2 |
| chr16_+_16888084 | 5.94 |
ENSMUST00000231514.2
|
Ypel1
|
yippee like 1 |
| chr6_-_53797748 | 5.89 |
ENSMUST00000127748.5
|
Tril
|
TLR4 interactor with leucine-rich repeats |
| chr11_-_55310724 | 5.87 |
ENSMUST00000108858.8
ENSMUST00000141530.2 |
Sparc
|
secreted acidic cysteine rich glycoprotein |
| chr9_+_59563872 | 5.84 |
ENSMUST00000215623.2
ENSMUST00000215660.2 ENSMUST00000217353.2 |
Pkm
|
pyruvate kinase, muscle |
| chr9_+_59563838 | 5.72 |
ENSMUST00000163694.4
|
Pkm
|
pyruvate kinase, muscle |
| chr11_+_110956980 | 5.53 |
ENSMUST00000042970.3
|
Kcnj2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr17_-_24752683 | 5.48 |
ENSMUST00000061764.14
|
Rab26
|
RAB26, member RAS oncogene family |
| chr11_-_107805830 | 5.46 |
ENSMUST00000039071.3
|
Cacng5
|
calcium channel, voltage-dependent, gamma subunit 5 |
| chr2_+_112097087 | 5.28 |
ENSMUST00000110987.9
ENSMUST00000028549.14 |
Slc12a6
|
solute carrier family 12, member 6 |
| chr10_-_109669053 | 5.22 |
ENSMUST00000238286.2
|
Nav3
|
neuron navigator 3 |
| chr10_+_116013256 | 5.12 |
ENSMUST00000155606.8
ENSMUST00000128399.2 |
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr14_+_119375753 | 5.05 |
ENSMUST00000065904.5
|
Hs6st3
|
heparan sulfate 6-O-sulfotransferase 3 |
| chr15_-_71599664 | 4.99 |
ENSMUST00000022953.10
|
Fam135b
|
family with sequence similarity 135, member B |
| chr15_+_34306812 | 4.96 |
ENSMUST00000226766.2
ENSMUST00000163455.9 ENSMUST00000022947.7 ENSMUST00000228570.2 ENSMUST00000227759.2 |
Matn2
|
matrilin 2 |
| chr12_-_11200306 | 4.85 |
ENSMUST00000055673.2
|
Kcns3
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
| chr12_+_99930757 | 4.80 |
ENSMUST00000160413.8
ENSMUST00000177549.8 ENSMUST00000049788.9 |
Kcnk13
|
potassium channel, subfamily K, member 13 |
| chr18_-_46444941 | 4.72 |
ENSMUST00000072835.7
|
Ccdc112
|
coiled-coil domain containing 112 |
| chr15_+_6451721 | 4.65 |
ENSMUST00000163082.2
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr6_-_23839419 | 4.51 |
ENSMUST00000115358.9
ENSMUST00000163871.9 |
Cadps2
|
Ca2+-dependent activator protein for secretion 2 |
| chr9_-_27066685 | 4.45 |
ENSMUST00000034472.16
|
Jam3
|
junction adhesion molecule 3 |
| chr8_-_85663976 | 4.36 |
ENSMUST00000109741.9
ENSMUST00000119820.2 |
Mast1
|
microtubule associated serine/threonine kinase 1 |
| chr3_-_122778052 | 4.26 |
ENSMUST00000199401.2
ENSMUST00000197314.5 ENSMUST00000197934.5 ENSMUST00000090379.7 |
Usp53
|
ubiquitin specific peptidase 53 |
| chr12_-_17374704 | 4.20 |
ENSMUST00000020884.16
ENSMUST00000095820.12 ENSMUST00000221129.2 ENSMUST00000127185.8 |
Atp6v1c2
|
ATPase, H+ transporting, lysosomal V1 subunit C2 |
| chr9_-_50650663 | 4.19 |
ENSMUST00000117093.2
ENSMUST00000121634.8 |
Dixdc1
|
DIX domain containing 1 |
| chr5_-_73805063 | 4.17 |
ENSMUST00000081170.9
|
Sgcb
|
sarcoglycan, beta (dystrophin-associated glycoprotein) |
| chr3_-_107993906 | 4.15 |
ENSMUST00000102638.8
ENSMUST00000102637.8 |
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr11_-_76514334 | 4.14 |
ENSMUST00000238684.2
|
Abr
|
active BCR-related gene |
| chr15_-_11907267 | 4.10 |
ENSMUST00000228489.2
|
Npr3
|
natriuretic peptide receptor 3 |
| chr14_+_55120875 | 4.08 |
ENSMUST00000134077.2
ENSMUST00000172844.8 ENSMUST00000133397.4 ENSMUST00000227108.2 |
Gm20521
Bcl2l2
|
predicted gene 20521 BCL2-like 2 |
| chr6_-_85742726 | 4.00 |
ENSMUST00000050780.8
|
Nat8f3
|
N-acetyltransferase 8 (GCN5-related) family member 3 |
| chr4_-_119395966 | 3.95 |
ENSMUST00000079611.13
|
Frg2f1
|
FSHD region gene 2 family member 1 |
| chrX_-_56549092 | 3.91 |
ENSMUST00000057645.6
|
Gpr101
|
G protein-coupled receptor 101 |
| chr13_-_98342659 | 3.90 |
ENSMUST00000109426.3
|
Arhgef28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr2_+_112096154 | 3.84 |
ENSMUST00000110991.9
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr7_+_126446588 | 3.82 |
ENSMUST00000141805.8
ENSMUST00000064110.14 ENSMUST00000205938.2 ENSMUST00000152051.8 |
Doc2a
|
double C2, alpha |
| chr10_+_116013122 | 3.81 |
ENSMUST00000148731.8
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr4_-_45084564 | 3.78 |
ENSMUST00000052236.13
|
Fbxo10
|
F-box protein 10 |
| chr15_+_98532624 | 3.73 |
ENSMUST00000003442.9
|
Cacnb3
|
calcium channel, voltage-dependent, beta 3 subunit |
| chr2_-_30014466 | 3.66 |
ENSMUST00000044751.14
|
Zer1
|
zyg-11 related, cell cycle regulator |
| chr17_-_26063391 | 3.63 |
ENSMUST00000176591.8
|
Rhot2
|
ras homolog family member T2 |
| chr17_-_26063488 | 3.62 |
ENSMUST00000176709.2
|
Rhot2
|
ras homolog family member T2 |
| chr4_-_135221926 | 3.55 |
ENSMUST00000102549.10
|
Nipal3
|
NIPA-like domain containing 3 |
| chr1_+_183078851 | 3.55 |
ENSMUST00000193625.2
|
Aida
|
axin interactor, dorsalization associated |
| chr16_+_44167484 | 3.54 |
ENSMUST00000050897.7
|
Spice1
|
spindle and centriole associated protein 1 |
| chr12_+_4642629 | 3.42 |
ENSMUST00000218402.2
|
Itsn2
|
intersectin 2 |
| chr6_-_90693471 | 3.40 |
ENSMUST00000101153.10
|
Iqsec1
|
IQ motif and Sec7 domain 1 |
| chr8_-_103512274 | 3.37 |
ENSMUST00000075190.5
|
Cdh11
|
cadherin 11 |
| chr18_-_3281752 | 3.34 |
ENSMUST00000140332.8
ENSMUST00000147138.8 |
Crem
|
cAMP responsive element modulator |
| chr2_-_126625343 | 3.17 |
ENSMUST00000028842.9
ENSMUST00000130356.3 |
Usp50
|
ubiquitin specific peptidase 50 |
| chr4_-_151129020 | 3.15 |
ENSMUST00000103204.11
|
Per3
|
period circadian clock 3 |
| chr3_+_37402795 | 3.13 |
ENSMUST00000200585.5
ENSMUST00000038885.10 |
Fgf2
|
fibroblast growth factor 2 |
| chr4_-_135221810 | 3.08 |
ENSMUST00000105856.9
|
Nipal3
|
NIPA-like domain containing 3 |
| chr2_-_30720345 | 2.90 |
ENSMUST00000041726.4
|
Asb6
|
ankyrin repeat and SOCS box-containing 6 |
| chr15_-_78602971 | 2.85 |
ENSMUST00000088592.6
|
Elfn2
|
leucine rich repeat and fibronectin type III, extracellular 2 |
| chr7_-_24211879 | 2.84 |
ENSMUST00000234781.2
|
Gm50092
|
predicted gene, 50092 |
| chr7_+_122977205 | 2.71 |
ENSMUST00000033025.7
ENSMUST00000206574.2 |
Lcmt1
|
leucine carboxyl methyltransferase 1 |
| chr7_-_127805518 | 2.57 |
ENSMUST00000033049.9
|
Cox6a2
|
cytochrome c oxidase subunit 6A2 |
| chr18_+_36693024 | 2.44 |
ENSMUST00000134146.8
|
Ankhd1
|
ankyrin repeat and KH domain containing 1 |
| chr16_+_16887991 | 2.41 |
ENSMUST00000232258.2
|
Ypel1
|
yippee like 1 |
| chr19_-_56996617 | 2.32 |
ENSMUST00000118800.8
ENSMUST00000111584.9 ENSMUST00000122359.8 ENSMUST00000148049.8 |
Afap1l2
|
actin filament associated protein 1-like 2 |
| chr16_+_3702493 | 2.30 |
ENSMUST00000176233.2
|
Gm20695
|
predicted gene 20695 |
| chr2_+_31840151 | 2.30 |
ENSMUST00000001920.13
ENSMUST00000151276.3 |
Aif1l
|
allograft inflammatory factor 1-like |
| chr19_+_47167554 | 2.28 |
ENSMUST00000235290.2
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr11_-_58346806 | 2.19 |
ENSMUST00000055204.6
|
Olfr30
|
olfactory receptor 30 |
| chrX_-_48877080 | 2.11 |
ENSMUST00000114893.8
|
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr3_-_57209357 | 2.00 |
ENSMUST00000196979.5
ENSMUST00000029376.13 |
Tm4sf1
|
transmembrane 4 superfamily member 1 |
| chr18_-_3281089 | 1.98 |
ENSMUST00000139537.2
ENSMUST00000124747.8 |
Crem
|
cAMP responsive element modulator |
| chr11_+_87483723 | 1.78 |
ENSMUST00000119628.8
|
Mtmr4
|
myotubularin related protein 4 |
| chr2_+_91087156 | 1.71 |
ENSMUST00000144394.8
ENSMUST00000028694.12 ENSMUST00000168916.8 ENSMUST00000156919.8 |
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr5_-_123822351 | 1.60 |
ENSMUST00000111564.8
ENSMUST00000063905.12 |
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr18_-_70186503 | 1.58 |
ENSMUST00000117692.8
ENSMUST00000069749.9 |
Rab27b
|
RAB27B, member RAS oncogene family |
| chr12_+_4642987 | 1.57 |
ENSMUST00000062580.8
ENSMUST00000220311.2 |
Itsn2
|
intersectin 2 |
| chr7_-_130931235 | 1.43 |
ENSMUST00000188899.2
|
Fam24b
|
family with sequence similarity 24 member B |
| chr17_-_40519480 | 1.41 |
ENSMUST00000033585.7
|
Pgk2
|
phosphoglycerate kinase 2 |
| chr5_-_137609691 | 1.33 |
ENSMUST00000031731.14
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr5_-_137609634 | 1.32 |
ENSMUST00000054564.13
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr12_+_105302853 | 1.26 |
ENSMUST00000180458.9
|
Tunar
|
Tcl1 upstream neural differentiation associated RNA |
| chr5_-_123822338 | 1.17 |
ENSMUST00000111561.8
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr19_-_61216834 | 1.14 |
ENSMUST00000076046.7
|
Csf2ra
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr9_-_21763898 | 1.12 |
ENSMUST00000217336.2
ENSMUST00000034728.9 |
Dock6
|
dedicator of cytokinesis 6 |
| chr19_+_4889394 | 1.12 |
ENSMUST00000088653.3
|
Ccdc87
|
coiled-coil domain containing 87 |
| chr7_+_106969950 | 1.11 |
ENSMUST00000073459.12
|
Syt9
|
synaptotagmin IX |
| chr7_-_119078472 | 1.08 |
ENSMUST00000209095.2
ENSMUST00000033263.6 ENSMUST00000207261.2 |
Umod
|
uromodulin |
| chr6_-_57885013 | 1.01 |
ENSMUST00000228076.2
ENSMUST00000228905.2 ENSMUST00000227650.2 ENSMUST00000227342.2 ENSMUST00000228257.2 |
Vmn1r22
|
vomeronasal 1 receptor 22 |
| chr10_-_129023288 | 0.98 |
ENSMUST00000072976.4
|
Olfr773
|
olfactory receptor 773 |
| chr18_-_3281727 | 0.97 |
ENSMUST00000154705.8
ENSMUST00000151084.8 |
Crem
|
cAMP responsive element modulator |
| chr3_-_124219673 | 0.90 |
ENSMUST00000029598.10
|
1700006A11Rik
|
RIKEN cDNA 1700006A11 gene |
| chr7_-_119078330 | 0.86 |
ENSMUST00000207460.2
|
Umod
|
uromodulin |
| chrX_-_151552457 | 0.85 |
ENSMUST00000101141.9
ENSMUST00000062317.5 |
Shroom2
|
shroom family member 2 |
| chr18_-_3280999 | 0.84 |
ENSMUST00000049942.13
|
Crem
|
cAMP responsive element modulator |
| chr7_-_127545896 | 0.80 |
ENSMUST00000118755.8
ENSMUST00000094026.10 |
Prss36
|
protease, serine 36 |
| chr7_-_134539993 | 0.76 |
ENSMUST00000171394.3
|
Insyn2a
|
inhibitory synaptic factor 2A |
| chr18_+_60880149 | 0.75 |
ENSMUST00000236652.2
ENSMUST00000235966.2 |
Rps14
|
ribosomal protein S14 |
| chr5_-_138169476 | 0.72 |
ENSMUST00000147920.2
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr10_-_19727165 | 0.69 |
ENSMUST00000059805.6
|
Slc35d3
|
solute carrier family 35, member D3 |
| chr7_-_29898236 | 0.66 |
ENSMUST00000001845.13
|
Capns1
|
calpain, small subunit 1 |
| chr16_+_3702604 | 0.62 |
ENSMUST00000115860.8
|
Naa60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr1_-_24139387 | 0.53 |
ENSMUST00000027337.15
|
Fam135a
|
family with sequence similarity 135, member A |
| chr9_+_91250864 | 0.51 |
ENSMUST00000173054.8
|
Zic4
|
zinc finger protein of the cerebellum 4 |
| chrX_+_84091915 | 0.48 |
ENSMUST00000239019.2
ENSMUST00000113992.3 ENSMUST00000113991.8 |
Dmd
|
dystrophin, muscular dystrophy |
| chr16_+_3702523 | 0.41 |
ENSMUST00000176625.8
ENSMUST00000186375.8 |
Naa60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr3_-_20209220 | 0.39 |
ENSMUST00000184552.2
|
Gyg
|
glycogenin |
| chr14_+_31750946 | 0.39 |
ENSMUST00000022460.11
|
Galnt15
|
polypeptide N-acetylgalactosaminyltransferase 15 |
| chr7_+_99659121 | 0.38 |
ENSMUST00000107084.8
|
Chrdl2
|
chordin-like 2 |
| chr13_-_81781238 | 0.36 |
ENSMUST00000126444.8
ENSMUST00000128585.9 ENSMUST00000146749.2 ENSMUST00000095585.11 |
Adgrv1
|
adhesion G protein-coupled receptor V1 |
| chr1_-_24139263 | 0.33 |
ENSMUST00000187369.7
ENSMUST00000187752.7 ENSMUST00000186999.7 |
Fam135a
|
family with sequence similarity 135, member A |
| chr10_+_116137277 | 0.28 |
ENSMUST00000092167.7
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr12_-_40087393 | 0.25 |
ENSMUST00000146905.2
|
Arl4a
|
ADP-ribosylation factor-like 4A |
| chr8_+_77628916 | 0.18 |
ENSMUST00000109912.8
ENSMUST00000128862.2 ENSMUST00000109911.8 |
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr7_+_126447080 | 0.15 |
ENSMUST00000147257.2
ENSMUST00000139174.2 |
Doc2a
|
double C2, alpha |
| chr4_-_41731142 | 0.13 |
ENSMUST00000171251.8
|
Arid3c
|
AT rich interactive domain 3C (BRIGHT-like) |
| chr9_-_22028370 | 0.12 |
ENSMUST00000213233.2
|
Elof1
|
ELF1 homolog, elongation factor 1 |
| chr2_-_168432111 | 0.11 |
ENSMUST00000029057.13
ENSMUST00000074618.10 |
Nfatc2
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2 |
| chr10_+_26105605 | 0.09 |
ENSMUST00000218301.2
ENSMUST00000164660.8 ENSMUST00000060716.6 |
Samd3
|
sterile alpha motif domain containing 3 |
| chr7_+_100742070 | 0.07 |
ENSMUST00000208439.2
|
Fchsd2
|
FCH and double SH3 domains 2 |
| chr5_+_89675288 | 0.02 |
ENSMUST00000048557.3
|
Npffr2
|
neuropeptide FF receptor 2 |
| chr10_+_56253531 | 0.02 |
ENSMUST00000220194.2
ENSMUST00000218834.2 |
Gja1
|
gap junction protein, alpha 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 24.0 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 3.6 | 10.9 | GO:0010232 | vascular transport(GO:0010232) milk ejection(GO:0060156) |
| 3.3 | 19.7 | GO:0007412 | axon target recognition(GO:0007412) |
| 3.1 | 12.2 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 2.4 | 12.2 | GO:0032901 | positive regulation of neurotrophin production(GO:0032901) |
| 2.3 | 9.1 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 1.8 | 5.5 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 1.8 | 10.7 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 1.7 | 5.0 | GO:0031104 | dendrite regeneration(GO:0031104) |
| 1.6 | 9.8 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 1.6 | 3.1 | GO:0021762 | substantia nigra development(GO:0021762) |
| 1.5 | 8.9 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 1.5 | 13.2 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 1.2 | 9.6 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 1.2 | 11.6 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 1.1 | 9.1 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 1.1 | 6.8 | GO:0015819 | lysine transport(GO:0015819) |
| 1.1 | 12.4 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 1.1 | 7.8 | GO:1901911 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 1.0 | 10.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 1.0 | 15.2 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.9 | 4.7 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.9 | 3.5 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.8 | 5.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.6 | 2.8 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.5 | 2.7 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.5 | 4.1 | GO:0060313 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.5 | 1.9 | GO:0072218 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.4 | 9.2 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.4 | 4.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.4 | 4.1 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.4 | 15.3 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.4 | 6.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.3 | 4.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.3 | 4.4 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.3 | 7.2 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.3 | 10.5 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 4.2 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.2 | 3.7 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.2 | 3.9 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.2 | 4.8 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 0.7 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.2 | 6.2 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.2 | 19.2 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.2 | 2.3 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.2 | 3.4 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.2 | 8.8 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.2 | 2.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 3.9 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.1 | 5.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 5.5 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 4.3 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 5.9 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.1 | 1.0 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 4.2 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.1 | 5.9 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.1 | 7.1 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.1 | 2.3 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.1 | 2.4 | GO:0032401 | establishment of melanosome localization(GO:0032401) |
| 0.1 | 3.2 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.1 | 4.0 | GO:0003401 | axis elongation(GO:0003401) |
| 0.1 | 2.3 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.1 | 1.1 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.1 | 1.0 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 3.7 | GO:0051438 | regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.1 | 6.0 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.1 | 1.3 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.2 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.0 | 0.4 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 2.8 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 4.6 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 3.8 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 2.6 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 1.4 | GO:0006096 | glycolytic process(GO:0006096) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.6 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 2.5 | 12.4 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 2.2 | 13.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 1.5 | 5.9 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 1.3 | 26.9 | GO:0031045 | dense core granule(GO:0031045) |
| 1.2 | 9.6 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 1.1 | 4.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.9 | 4.7 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.8 | 10.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.7 | 5.9 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.5 | 10.9 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.5 | 4.4 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.3 | 3.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 2.8 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.2 | 3.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 3.7 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.2 | 7.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 1.6 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.2 | 13.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 2.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 5.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 10.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 12.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 5.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.4 | GO:0002142 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.1 | 22.2 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.1 | 5.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 2.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 6.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 2.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 1.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 4.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 10.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 6.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 3.0 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 9.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 3.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 3.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 9.1 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 4.0 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 28.2 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.5 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 6.7 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 0.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 7.0 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 19.7 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 2.6 | 13.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 2.3 | 11.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 2.3 | 9.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 1.8 | 5.5 | GO:0019002 | GMP binding(GO:0019002) |
| 1.6 | 9.6 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 1.6 | 10.9 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 1.4 | 6.8 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 1.3 | 5.0 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 1.1 | 7.8 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 1.0 | 6.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.9 | 12.2 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.9 | 6.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.7 | 4.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.6 | 2.3 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 0.5 | 2.7 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.4 | 18.3 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.4 | 12.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.4 | 4.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.4 | 10.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.4 | 1.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.4 | 5.5 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.4 | 3.9 | GO:0004935 | adrenergic receptor activity(GO:0004935) |
| 0.3 | 6.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.3 | 1.4 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.2 | 2.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 4.8 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.2 | 18.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 4.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 4.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 16.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 13.6 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.2 | 5.9 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.2 | 5.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.2 | 1.9 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 3.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 5.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 9.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 8.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 8.1 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 2.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 2.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 4.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 2.6 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 8.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 13.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 3.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.7 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 11.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 1.0 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 0.7 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.2 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.1 | 1.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 4.9 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 19.9 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 2.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 9.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.5 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 5.9 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 4.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 5.5 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 2.2 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 19.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 10.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.2 | 10.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 14.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 15.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 5.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 4.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 6.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 6.9 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 4.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 3.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 10.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 13.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 4.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 4.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 15.5 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.5 | 12.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.3 | 4.8 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 3.7 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.2 | 4.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 3.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 9.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 4.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 6.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 5.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 4.9 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.1 | 3.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 8.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 20.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 4.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 2.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.1 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |