Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Six3_Six1_Six2
Z-value: 0.61
Transcription factors associated with Six3_Six1_Six2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
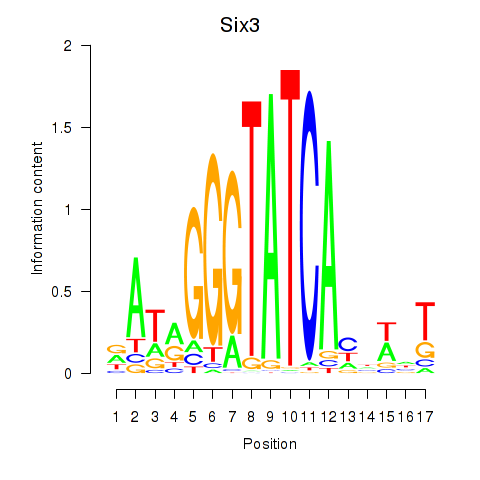
Six3
|
ENSMUSG00000038805.11 | Six3 |
|
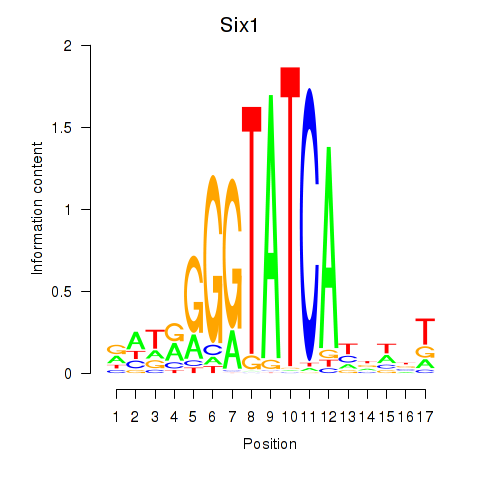
Six1
|
ENSMUSG00000051367.9 | Six1 |
|
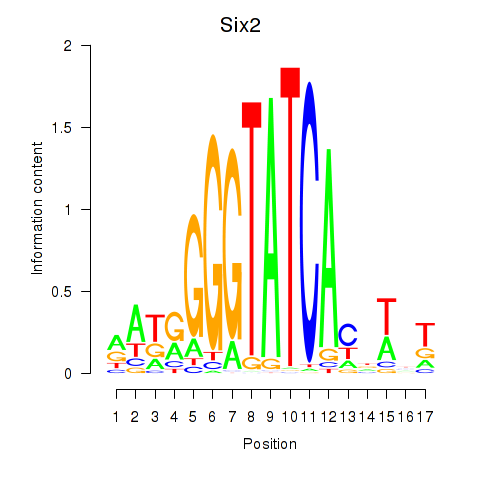
Six2
|
ENSMUSG00000024134.12 | Six2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Six1 | mm39_v1_chr12_-_73093953_73093953 | -0.29 | 1.4e-02 | Click! |
| Six3 | mm39_v1_chr17_+_85920860_85921036 | -0.21 | 8.2e-02 | Click! |
| Six2 | mm39_v1_chr17_-_85995680_85995703 | -0.17 | 1.4e-01 | Click! |
Activity profile of Six3_Six1_Six2 motif
Sorted Z-values of Six3_Six1_Six2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Six3_Six1_Six2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_65162267 | 5.38 |
ENSMUST00000050047.4
ENSMUST00000148020.8 |
D630023F18Rik
|
RIKEN cDNA D630023F18 gene |
| chr7_-_79765042 | 3.29 |
ENSMUST00000206714.2
ENSMUST00000107384.10 |
Idh2
|
isocitrate dehydrogenase 2 (NADP+), mitochondrial |
| chr9_+_32305259 | 3.28 |
ENSMUST00000172015.3
|
Kcnj1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr14_+_75479727 | 3.26 |
ENSMUST00000022576.10
|
Cpb2
|
carboxypeptidase B2 (plasma) |
| chr9_+_98305014 | 2.71 |
ENSMUST00000052068.11
|
Rbp1
|
retinol binding protein 1, cellular |
| chr2_+_106523532 | 2.62 |
ENSMUST00000111063.8
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr16_+_22738987 | 2.43 |
ENSMUST00000023587.12
|
Fetub
|
fetuin beta |
| chr5_+_122239830 | 2.26 |
ENSMUST00000146733.5
|
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr15_+_98468885 | 2.25 |
ENSMUST00000023728.8
|
Tex49
|
testis expressed 49 |
| chr15_-_75766321 | 2.04 |
ENSMUST00000023237.8
|
Naprt
|
nicotinate phosphoribosyltransferase |
| chr3_+_145926709 | 1.96 |
ENSMUST00000039164.4
|
Lpar3
|
lysophosphatidic acid receptor 3 |
| chr10_-_128016135 | 1.91 |
ENSMUST00000238843.2
ENSMUST00000099139.9 |
Rbms2
|
RNA binding motif, single stranded interacting protein 2 |
| chrX_-_141749704 | 1.89 |
ENSMUST00000041317.3
|
Ammecr1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr7_+_30676465 | 1.84 |
ENSMUST00000058093.6
|
Fam187b
|
family with sequence similarity 187, member B |
| chr7_+_15484302 | 1.77 |
ENSMUST00000098802.10
ENSMUST00000173053.2 |
Obox5
|
oocyte specific homeobox 5 |
| chr3_-_92734546 | 1.76 |
ENSMUST00000072363.5
|
Kprp
|
keratinocyte expressed, proline-rich |
| chr2_-_152382014 | 1.35 |
ENSMUST00000060598.4
|
Defb29
|
defensin beta 29 |
| chr7_+_15484056 | 1.25 |
ENSMUST00000173455.8
|
Obox5
|
oocyte specific homeobox 5 |
| chr16_+_22739191 | 1.17 |
ENSMUST00000116625.10
|
Fetub
|
fetuin beta |
| chr16_+_22739028 | 1.17 |
ENSMUST00000232097.2
|
Fetub
|
fetuin beta |
| chr2_-_88534814 | 0.94 |
ENSMUST00000216928.2
ENSMUST00000216977.2 |
Olfr1196
|
olfactory receptor 1196 |
| chr5_+_23992689 | 0.90 |
ENSMUST00000120869.6
ENSMUST00000030852.13 ENSMUST00000117783.8 ENSMUST00000115113.3 |
Rint1
|
RAD50 interactor 1 |
| chr7_-_5119910 | 0.89 |
ENSMUST00000179971.8
ENSMUST00000045215.9 ENSMUST00000208468.2 |
Rfpl4
|
ret finger protein-like 4 |
| chr5_+_21748523 | 0.89 |
ENSMUST00000035651.6
|
Lrrc17
|
leucine rich repeat containing 17 |
| chr5_+_139408906 | 0.88 |
ENSMUST00000066211.5
|
Gper1
|
G protein-coupled estrogen receptor 1 |
| chr3_+_82915031 | 0.86 |
ENSMUST00000048486.13
ENSMUST00000194175.2 |
Fgg
|
fibrinogen gamma chain |
| chr19_+_27194757 | 0.85 |
ENSMUST00000047645.13
ENSMUST00000167487.8 |
Vldlr
|
very low density lipoprotein receptor |
| chr7_+_14393658 | 0.82 |
ENSMUST00000069740.14
ENSMUST00000183424.2 |
Obox7
|
oocyte specific homeobox 7 |
| chr7_+_43339842 | 0.79 |
ENSMUST00000056329.7
|
Klk14
|
kallikrein related-peptidase 14 |
| chr9_+_110856425 | 0.77 |
ENSMUST00000199313.2
|
Ltf
|
lactotransferrin |
| chr7_+_14393421 | 0.76 |
ENSMUST00000183788.8
|
Obox7
|
oocyte specific homeobox 7 |
| chr4_-_116508842 | 0.72 |
ENSMUST00000030455.15
|
Akr1a1
|
aldo-keto reductase family 1, member A1 (aldehyde reductase) |
| chr6_+_38511758 | 0.67 |
ENSMUST00000019833.5
|
Fmc1
|
formation of mitochondrial complex V assembly factor 1 |
| chr9_+_38725910 | 0.67 |
ENSMUST00000213164.2
|
Olfr922
|
olfactory receptor 922 |
| chrX_-_73416869 | 0.66 |
ENSMUST00000073067.11
ENSMUST00000037967.6 |
Slc10a3
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 3 |
| chr17_-_57024660 | 0.62 |
ENSMUST00000164907.3
ENSMUST00000233208.2 |
Vmac
|
vimentin-type intermediate filament associated coiled-coil protein |
| chr19_-_38032006 | 0.60 |
ENSMUST00000172095.3
ENSMUST00000041475.16 |
Myof
|
myoferlin |
| chr19_-_38031774 | 0.56 |
ENSMUST00000226068.2
|
Myof
|
myoferlin |
| chr17_-_57023788 | 0.54 |
ENSMUST00000067931.7
|
Vmac
|
vimentin-type intermediate filament associated coiled-coil protein |
| chr4_-_60457902 | 0.53 |
ENSMUST00000084548.11
ENSMUST00000103012.10 ENSMUST00000107499.4 |
Mup1
|
major urinary protein 1 |
| chr7_+_26250237 | 0.52 |
ENSMUST00000122040.7
ENSMUST00000117252.4 |
Nlrp9a
|
NLR family, pyrin domain containing 9A |
| chr17_-_41225241 | 0.51 |
ENSMUST00000166343.3
|
Glyatl3
|
glycine-N-acyltransferase-like 3 |
| chr17_+_57024759 | 0.51 |
ENSMUST00000002452.8
ENSMUST00000233832.2 ENSMUST00000233233.2 |
Ndufa11
|
NADH:ubiquinone oxidoreductase subunit A11 |
| chr5_+_138805835 | 0.49 |
ENSMUST00000179205.2
|
Foxl3
|
forkhead box L3 |
| chr8_+_95720864 | 0.49 |
ENSMUST00000212141.2
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr13_+_94219934 | 0.48 |
ENSMUST00000156071.2
|
Lhfpl2
|
lipoma HMGIC fusion partner-like 2 |
| chr14_+_58035614 | 0.47 |
ENSMUST00000128764.9
|
Sap18
|
Sin3-associated polypeptide 18 |
| chr9_+_103879745 | 0.47 |
ENSMUST00000035167.15
ENSMUST00000194774.6 |
Nphp3
|
nephronophthisis 3 (adolescent) |
| chr11_+_58707754 | 0.46 |
ENSMUST00000082220.5
|
Olfr313
|
olfactory receptor 313 |
| chr19_-_55304392 | 0.45 |
ENSMUST00000224291.2
ENSMUST00000225495.2 ENSMUST00000076891.7 ENSMUST00000224897.2 |
Zdhhc6
|
zinc finger, DHHC domain containing 6 |
| chr14_+_58035640 | 0.44 |
ENSMUST00000111269.2
|
Sap18
|
Sin3-associated polypeptide 18 |
| chr7_+_6470100 | 0.43 |
ENSMUST00000207658.4
ENSMUST00000215302.3 |
Olfr1346
|
olfactory receptor 1346 |
| chr5_-_86780277 | 0.43 |
ENSMUST00000116553.9
|
Tmprss11f
|
transmembrane protease, serine 11f |
| chr13_-_40882417 | 0.42 |
ENSMUST00000225180.2
|
Tfap2a
|
transcription factor AP-2, alpha |
| chr4_-_106397733 | 0.42 |
ENSMUST00000238822.2
ENSMUST00000239147.2 |
Gm53010
Tmem61
|
predicted gene, 53010 transmembrane protein 61 |
| chr4_-_144291704 | 0.41 |
ENSMUST00000105748.2
|
Aadacl4fm2
|
AADACL4 family member 2 |
| chr7_+_43837629 | 0.41 |
ENSMUST00000073713.8
|
Klk1b24
|
kallikrein 1-related peptidase b24 |
| chr6_-_52195663 | 0.41 |
ENSMUST00000134367.4
|
Hoxa7
|
homeobox A7 |
| chr7_-_12819142 | 0.41 |
ENSMUST00000094829.2
|
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr9_-_121745354 | 0.40 |
ENSMUST00000062474.5
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chr9_+_119170486 | 0.39 |
ENSMUST00000175743.8
ENSMUST00000176397.8 |
Acaa1a
|
acetyl-Coenzyme A acyltransferase 1A |
| chr7_+_29931735 | 0.39 |
ENSMUST00000108193.2
ENSMUST00000108192.2 |
Polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chr11_-_8989582 | 0.37 |
ENSMUST00000043377.6
|
Sun3
|
Sad1 and UNC84 domain containing 3 |
| chr6_-_58449863 | 0.35 |
ENSMUST00000226390.2
|
Vmn1r31
|
vomeronasal 1 receptor 31 |
| chr6_-_119925387 | 0.35 |
ENSMUST00000162541.8
|
Wnk1
|
WNK lysine deficient protein kinase 1 |
| chr19_+_55304703 | 0.34 |
ENSMUST00000225529.2
ENSMUST00000223690.2 ENSMUST00000095950.3 |
Vti1a
|
vesicle transport through interaction with t-SNAREs 1A |
| chr17_+_25352353 | 0.33 |
ENSMUST00000162862.3
ENSMUST00000040729.9 |
Clcn7
|
chloride channel, voltage-sensitive 7 |
| chr9_-_36708569 | 0.30 |
ENSMUST00000163192.11
|
Ei24
|
etoposide induced 2.4 mRNA |
| chr9_+_65048454 | 0.30 |
ENSMUST00000034961.6
ENSMUST00000217371.2 |
Igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr10_+_77891161 | 0.29 |
ENSMUST00000131825.8
ENSMUST00000139539.8 ENSMUST00000123940.2 |
Dnmt3l
|
DNA (cytosine-5-)-methyltransferase 3-like |
| chr6_+_48514578 | 0.29 |
ENSMUST00000203011.2
|
Atp6v0e2
|
ATPase, H+ transporting, lysosomal V0 subunit E2 |
| chr14_+_64229914 | 0.28 |
ENSMUST00000058229.6
|
Rp1l1
|
retinitis pigmentosa 1 homolog like 1 |
| chr7_-_126275529 | 0.27 |
ENSMUST00000106372.11
ENSMUST00000155419.3 ENSMUST00000106373.9 |
Sult1a1
|
sulfotransferase family 1A, phenol-preferring, member 1 |
| chr11_-_69586347 | 0.27 |
ENSMUST00000181261.2
|
Tnfsf12
|
tumor necrosis factor (ligand) superfamily, member 12 |
| chr6_+_48514518 | 0.26 |
ENSMUST00000040361.8
|
Atp6v0e2
|
ATPase, H+ transporting, lysosomal V0 subunit E2 |
| chr6_-_115652831 | 0.26 |
ENSMUST00000112949.8
|
Raf1
|
v-raf-leukemia viral oncogene 1 |
| chr9_+_119170360 | 0.25 |
ENSMUST00000039784.12
|
Acaa1a
|
acetyl-Coenzyme A acyltransferase 1A |
| chr7_-_48149069 | 0.25 |
ENSMUST00000098433.5
|
Mrgprx2
|
MAS-related GPR, member X2 |
| chr17_-_56312555 | 0.24 |
ENSMUST00000043785.8
|
Stap2
|
signal transducing adaptor family member 2 |
| chr1_-_83020201 | 0.24 |
ENSMUST00000188323.2
|
Krtap28-10
|
keratin associated protein 28-10 |
| chr1_+_44158111 | 0.23 |
ENSMUST00000155917.8
|
Bivm
|
basic, immunoglobulin-like variable motif containing |
| chr2_-_13276205 | 0.22 |
ENSMUST00000191959.6
ENSMUST00000028059.9 |
Rsu1
|
Ras suppressor protein 1 |
| chr9_-_36708599 | 0.22 |
ENSMUST00000238932.2
ENSMUST00000115086.13 |
Ei24
|
etoposide induced 2.4 mRNA |
| chr8_+_85558151 | 0.22 |
ENSMUST00000036734.6
|
Gadd45gip1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
| chr7_-_29931612 | 0.22 |
ENSMUST00000006254.6
|
Tbcb
|
tubulin folding cofactor B |
| chr9_+_38686470 | 0.21 |
ENSMUST00000071681.4
|
Olfr921
|
olfactory receptor 921 |
| chrX_+_37861548 | 0.21 |
ENSMUST00000050744.6
|
6030498E09Rik
|
RIKEN cDNA 6030498E09 gene |
| chr2_-_13276074 | 0.20 |
ENSMUST00000137670.3
ENSMUST00000114791.9 |
Rsu1
|
Ras suppressor protein 1 |
| chr11_+_49355873 | 0.20 |
ENSMUST00000213674.2
ENSMUST00000204518.3 |
Olfr1386
|
olfactory receptor 1386 |
| chrX_-_73416824 | 0.20 |
ENSMUST00000178691.2
ENSMUST00000114146.8 |
Ubl4a
Slc10a3
|
ubiquitin-like 4A solute carrier family 10 (sodium/bile acid cotransporter family), member 3 |
| chr14_-_75830550 | 0.19 |
ENSMUST00000164082.9
ENSMUST00000169658.9 |
Cby2
|
chibby family member 2 |
| chr1_+_40305738 | 0.18 |
ENSMUST00000114795.3
|
Il1r1
|
interleukin 1 receptor, type I |
| chr9_+_123921573 | 0.18 |
ENSMUST00000111442.3
ENSMUST00000171499.3 |
Ccr5
|
chemokine (C-C motif) receptor 5 |
| chr7_-_103393740 | 0.18 |
ENSMUST00000216300.2
|
Olfr629
|
olfactory receptor 629 |
| chr2_+_87725306 | 0.17 |
ENSMUST00000217436.2
|
Olfr1153
|
olfactory receptor 1153 |
| chr16_-_18052937 | 0.17 |
ENSMUST00000076957.7
|
Zdhhc8
|
zinc finger, DHHC domain containing 8 |
| chr1_+_118104272 | 0.17 |
ENSMUST00000186264.2
|
Gm29106
|
predicted gene 29106 |
| chr11_+_120612369 | 0.17 |
ENSMUST00000142229.2
|
Rac3
|
Rac family small GTPase 3 |
| chr7_+_29931309 | 0.17 |
ENSMUST00000019882.16
ENSMUST00000149654.8 |
Polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chr8_-_87611849 | 0.17 |
ENSMUST00000034074.8
|
N4bp1
|
NEDD4 binding protein 1 |
| chr7_-_102507962 | 0.15 |
ENSMUST00000213481.2
ENSMUST00000209952.2 |
Olfr566
|
olfactory receptor 566 |
| chr1_+_92516054 | 0.15 |
ENSMUST00000062964.5
|
Olfr1412
|
olfactory receptor 1412 |
| chr9_-_35701371 | 0.15 |
ENSMUST00000034619.2
|
Pate6
|
prostate and testis expressed 6 |
| chrX_-_110446022 | 0.14 |
ENSMUST00000156639.2
|
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr16_-_16491881 | 0.14 |
ENSMUST00000057886.6
|
Olfr19
|
olfactory receptor 19 |
| chr1_+_173959083 | 0.14 |
ENSMUST00000214751.2
|
Olfr424
|
olfactory receptor 424 |
| chr2_-_85917726 | 0.13 |
ENSMUST00000216886.2
ENSMUST00000213333.2 ENSMUST00000216020.2 |
Olfr1037
|
olfactory receptor 1037 |
| chr9_-_39920878 | 0.13 |
ENSMUST00000216463.3
|
Olfr980
|
olfactory receptor 980 |
| chr3_-_88332401 | 0.13 |
ENSMUST00000168755.7
ENSMUST00000193433.6 ENSMUST00000195657.6 ENSMUST00000057935.9 |
Slc25a44
|
solute carrier family 25, member 44 |
| chr16_-_23751144 | 0.12 |
ENSMUST00000231038.2
|
Rtp2
|
receptor transporter protein 2 |
| chr2_-_111320501 | 0.11 |
ENSMUST00000099616.2
|
Olfr1290
|
olfactory receptor 1290 |
| chr11_-_73800125 | 0.11 |
ENSMUST00000215690.2
|
Olfr395
|
olfactory receptor 395 |
| chr11_+_120612278 | 0.11 |
ENSMUST00000018156.12
|
Rac3
|
Rac family small GTPase 3 |
| chr6_-_120893725 | 0.10 |
ENSMUST00000145948.2
|
Bid
|
BH3 interacting domain death agonist |
| chr7_-_103420801 | 0.10 |
ENSMUST00000106878.3
|
Olfr69
|
olfactory receptor 69 |
| chr19_+_46329552 | 0.09 |
ENSMUST00000128041.8
|
Mfsd13a
|
major facilitator superfamily domain containing 13a |
| chr3_+_32760447 | 0.08 |
ENSMUST00000194781.6
|
Actl6a
|
actin-like 6A |
| chr7_-_102368277 | 0.08 |
ENSMUST00000216312.2
|
Olfr33
|
olfactory receptor 33 |
| chr16_-_58647137 | 0.07 |
ENSMUST00000215069.2
ENSMUST00000079955.6 |
Olfr175
|
olfactory receptor 175 |
| chr7_+_102965522 | 0.07 |
ENSMUST00000216456.2
|
Olfr597
|
olfactory receptor 597 |
| chr6_+_17743581 | 0.07 |
ENSMUST00000000674.13
ENSMUST00000077080.9 |
St7
|
suppression of tumorigenicity 7 |
| chr4_-_106588122 | 0.06 |
ENSMUST00000148281.2
|
Mroh7
|
maestro heat-like repeat family member 7 |
| chr15_+_38662158 | 0.06 |
ENSMUST00000022904.8
ENSMUST00000228820.2 |
Atp6v1c1
|
ATPase, H+ transporting, lysosomal V1 subunit C1 |
| chrX_+_30768610 | 0.05 |
ENSMUST00000179532.2
|
Btbd35f29
|
BTB domain containing 35, family member 29 |
| chr6_-_57306479 | 0.05 |
ENSMUST00000227283.2
ENSMUST00000228356.2 |
Vmn1r16
|
vomeronasal 1 receptor 16 |
| chr6_-_120893771 | 0.05 |
ENSMUST00000004560.12
|
Bid
|
BH3 interacting domain death agonist |
| chrX_+_23748792 | 0.04 |
ENSMUST00000116614.3
|
Tesl1
|
testin LIM domain protein like 1 |
| chr10_-_12744025 | 0.04 |
ENSMUST00000219660.2
|
Utrn
|
utrophin |
| chr13_-_74440402 | 0.04 |
ENSMUST00000109640.8
ENSMUST00000022059.14 |
Ahrr
|
aryl-hydrocarbon receptor repressor |
| chr2_-_76698725 | 0.04 |
ENSMUST00000149616.8
ENSMUST00000152185.8 ENSMUST00000130915.8 ENSMUST00000155365.8 ENSMUST00000128071.8 |
Ttn
|
titin |
| chr15_-_103218876 | 0.03 |
ENSMUST00000079824.6
|
Gpr84
|
G protein-coupled receptor 84 |
| chr17_+_56312672 | 0.03 |
ENSMUST00000133998.8
|
Mpnd
|
MPN domain containing |
| chrX_-_31034822 | 0.03 |
ENSMUST00000238426.2
|
Btbd35f19
|
BTB domain containing 35, family member 19 |
| chrX_-_33139812 | 0.03 |
ENSMUST00000105117.3
|
Btbd35f14
|
BTB domain containing 35, family member 14 |
| chrX_+_33094635 | 0.03 |
ENSMUST00000177912.2
|
Btbd35f13
|
BTB domain containing 35, family member 13 |
| chrX_-_33014777 | 0.03 |
ENSMUST00000186329.2
|
Btbd35f15
|
BTB domain containing 35, family member 15 |
| chrY_-_40731110 | 0.03 |
ENSMUST00000177579.2
|
Gm21797
|
predicted gene, 21797 |
| chrX_+_4703838 | 0.02 |
ENSMUST00000105011.4
|
Btbd35f4
|
BTB domain containing 35, family member 4 |
| chrY_+_41361843 | 0.02 |
ENSMUST00000178559.2
|
Gm21800
|
predicted gene, 21800 |
| chrX_+_31608531 | 0.02 |
ENSMUST00000238528.2
|
Btbd35f22
|
BTB domain containing 35, family member 22 |
| chrX_+_32750842 | 0.02 |
ENSMUST00000178827.3
|
Btbd35f12
|
BTB domain containing 35, family member 12 |
| chr19_+_9824919 | 0.02 |
ENSMUST00000179814.3
|
Scgb2a2
|
secretoglobin, family 2A, member 2 |
| chrX_+_32411401 | 0.01 |
ENSMUST00000178747.3
|
Btbd35f5
|
BTB domain containing 35, family member 5 |
| chrX_-_33394003 | 0.01 |
ENSMUST00000179466.2
|
Btbd35f6
|
BTB domain containing 35, family member 6 |
| chrY_+_13941738 | 0.01 |
ENSMUST00000178689.2
|
Gm21767
|
predicted gene, 21767 |
| chrY_+_18654482 | 0.01 |
ENSMUST00000179302.2
|
Gm21822
|
predicted gene, 21822 |
| chrX_+_31839202 | 0.01 |
ENSMUST00000179991.3
|
Btbd35f2
|
BTB domain containing 35, family member 2 |
| chr16_-_11021116 | 0.01 |
ENSMUST00000230002.2
ENSMUST00000119953.2 |
Rsl1d1
|
ribosomal L1 domain containing 1 |
| chr8_+_72704882 | 0.01 |
ENSMUST00000131237.8
ENSMUST00000136516.9 ENSMUST00000109997.10 ENSMUST00000132848.2 |
Zfp961
|
zinc finger protein 961 |
| chrX_+_168468186 | 0.01 |
ENSMUST00000112107.8
ENSMUST00000112104.8 |
Mid1
|
midline 1 |
| chrY_-_13465579 | 0.01 |
ENSMUST00000178428.2
|
Gm21866
|
predicted gene, 21866 |
| chrX_-_153673263 | 0.01 |
ENSMUST00000096852.5
|
Cldn34b1
|
claudin 34B1 |
| chr9_+_72866067 | 0.01 |
ENSMUST00000098567.9
ENSMUST00000034734.9 |
Dnaaf4
|
dynein axonemal assembly factor 4 |
| chr14_+_123897383 | 0.00 |
ENSMUST00000049681.14
|
Itgbl1
|
integrin, beta-like 1 |
| chrX_+_32070863 | 0.00 |
ENSMUST00000238237.2
|
Btbd35f1
|
BTB domain containing 35, family member 1 |
| chrX_-_90826588 | 0.00 |
ENSMUST00000082048.3
|
Mageb5
|
MAGE family member B5 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:1904464 | regulation of matrix metallopeptidase secretion(GO:1904464) matrix metallopeptidase secretion(GO:1990773) |
| 1.1 | 3.3 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.9 | 2.7 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.8 | 2.3 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.4 | 3.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.3 | 0.9 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.3 | 0.9 | GO:2000724 | response to mineralocorticoid(GO:0051385) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.2 | 0.8 | GO:1900190 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.1 | 2.0 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 0.7 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.1 | 0.9 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.1 | 0.9 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 2.0 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.3 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.1 | 4.8 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 0.4 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.1 | 0.4 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.1 | 0.5 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) maintenance of organ identity(GO:0048496) |
| 0.1 | 1.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.9 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 0.5 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.1 | 0.6 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.8 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.2 | GO:0010286 | heat acclimation(GO:0010286) |
| 0.0 | 0.5 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.2 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.3 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.3 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.0 | 0.4 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.3 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.0 | 0.4 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.2 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.2 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.5 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.7 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.0 | 0.6 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.2 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.6 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.0 | 0.3 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.5 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 0.8 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.1 | 2.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.5 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 1.2 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.9 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.5 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.6 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.4 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.9 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.2 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 3.9 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.7 | 2.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.4 | 3.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.3 | 0.9 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.3 | 0.9 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.3 | 2.0 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 4.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 0.6 | GO:0008775 | acetate CoA-transferase activity(GO:0008775) |
| 0.2 | 0.5 | GO:0005009 | insulin-activated receptor activity(GO:0005009) |
| 0.2 | 2.7 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 0.9 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 3.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.7 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.5 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 1.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.2 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 2.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.8 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.6 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.6 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 2.7 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 2.0 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 0.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 3.3 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 2.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.6 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.3 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |