Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Six4
Z-value: 0.76
Transcription factors associated with Six4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Six4
|
ENSMUSG00000034460.10 | Six4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Six4 | mm39_v1_chr12_-_73160181_73160230 | -0.40 | 4.2e-04 | Click! |
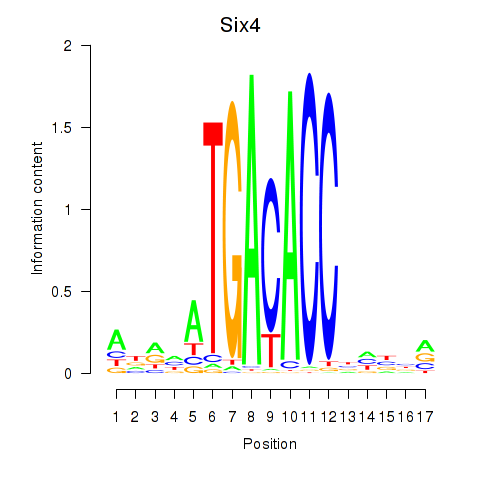
Activity profile of Six4 motif
Sorted Z-values of Six4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Six4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_78137927 | 6.87 |
ENSMUST00000098537.4
|
Gsta1
|
glutathione S-transferase, alpha 1 (Ya) |
| chr9_+_78197205 | 6.17 |
ENSMUST00000119823.8
ENSMUST00000121273.2 |
Gsta5
|
glutathione S-transferase alpha 5 |
| chr12_-_57592907 | 5.29 |
ENSMUST00000044380.8
|
Foxa1
|
forkhead box A1 |
| chr13_+_93810911 | 5.24 |
ENSMUST00000048001.8
|
Dmgdh
|
dimethylglycine dehydrogenase precursor |
| chr3_+_40905066 | 4.60 |
ENSMUST00000191805.7
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr2_+_24970327 | 4.58 |
ENSMUST00000044078.10
ENSMUST00000114380.9 |
Entpd8
|
ectonucleoside triphosphate diphosphohydrolase 8 |
| chr13_-_93810808 | 4.42 |
ENSMUST00000015941.8
|
Bhmt2
|
betaine-homocysteine methyltransferase 2 |
| chr7_+_26819334 | 4.36 |
ENSMUST00000003100.10
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2 |
| chr3_+_40905216 | 4.14 |
ENSMUST00000191872.6
ENSMUST00000200432.2 |
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr3_+_92864693 | 3.85 |
ENSMUST00000059053.11
|
Lce3d
|
late cornified envelope 3D |
| chr17_+_48717007 | 2.44 |
ENSMUST00000167180.8
ENSMUST00000046651.7 ENSMUST00000233426.2 |
Oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr13_-_72111832 | 2.17 |
ENSMUST00000077337.9
|
Irx1
|
Iroquois homeobox 1 |
| chr17_-_25564501 | 2.15 |
ENSMUST00000153118.2
ENSMUST00000146856.3 |
Tpsab1
|
tryptase alpha/beta 1 |
| chr10_+_75399920 | 2.15 |
ENSMUST00000141062.8
ENSMUST00000152657.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr18_+_60515755 | 2.06 |
ENSMUST00000237185.2
|
Iigp1
|
interferon inducible GTPase 1 |
| chr18_+_60345152 | 2.05 |
ENSMUST00000031549.6
|
Gm4951
|
predicted gene 4951 |
| chr3_-_75864195 | 2.00 |
ENSMUST00000038563.14
ENSMUST00000167078.8 ENSMUST00000117242.8 |
Golim4
|
golgi integral membrane protein 4 |
| chr15_-_103163860 | 1.97 |
ENSMUST00000075192.13
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr2_+_31204314 | 1.94 |
ENSMUST00000113532.9
|
Hmcn2
|
hemicentin 2 |
| chr9_+_30941924 | 1.84 |
ENSMUST00000216649.2
ENSMUST00000115222.10 |
Zbtb44
|
zinc finger and BTB domain containing 44 |
| chr6_+_35229628 | 1.77 |
ENSMUST00000130875.8
|
1810058I24Rik
|
RIKEN cDNA 1810058I24 gene |
| chr13_+_51799268 | 1.65 |
ENSMUST00000075853.6
|
Cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr10_-_3217767 | 1.62 |
ENSMUST00000170893.3
|
H60c
|
histocompatibility 60c |
| chr4_-_133746138 | 1.60 |
ENSMUST00000051674.3
|
Lin28a
|
lin-28 homolog A (C. elegans) |
| chr14_-_56322654 | 1.57 |
ENSMUST00000015594.9
|
Mcpt8
|
mast cell protease 8 |
| chr11_+_3939924 | 1.52 |
ENSMUST00000109981.2
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr10_-_3217718 | 1.47 |
ENSMUST00000216211.2
|
H60c
|
histocompatibility 60c |
| chr13_+_54225828 | 1.40 |
ENSMUST00000021930.10
|
Sfxn1
|
sideroflexin 1 |
| chr7_+_37883300 | 1.38 |
ENSMUST00000179992.10
|
1600014C10Rik
|
RIKEN cDNA 1600014C10 gene |
| chr5_-_41921834 | 1.32 |
ENSMUST00000060820.8
|
Nkx3-2
|
NK3 homeobox 2 |
| chr19_-_6957789 | 1.32 |
ENSMUST00000070878.9
ENSMUST00000177752.9 |
Fkbp2
|
FK506 binding protein 2 |
| chr11_+_49135018 | 1.29 |
ENSMUST00000167400.8
ENSMUST00000081794.7 |
Mgat1
|
mannoside acetylglucosaminyltransferase 1 |
| chr10_+_127478844 | 1.28 |
ENSMUST00000092074.12
ENSMUST00000120279.2 |
Stat6
|
signal transducer and activator of transcription 6 |
| chr5_-_132570710 | 1.21 |
ENSMUST00000182974.9
|
Auts2
|
autism susceptibility candidate 2 |
| chr5_-_145128321 | 1.21 |
ENSMUST00000161845.2
|
Atp5j2
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F2 |
| chr3_+_138058139 | 1.13 |
ENSMUST00000090166.5
|
Adh6b
|
alcohol dehydrogenase 6B (class V) |
| chr18_-_70186503 | 1.12 |
ENSMUST00000117692.8
ENSMUST00000069749.9 |
Rab27b
|
RAB27B, member RAS oncogene family |
| chr2_-_89774457 | 1.12 |
ENSMUST00000090695.3
|
Olfr1259
|
olfactory receptor 1259 |
| chr10_-_79481565 | 1.07 |
ENSMUST00000095464.3
|
Odf3l2
|
outer dense fiber of sperm tails 3-like 2 |
| chr4_-_144513121 | 1.02 |
ENSMUST00000105746.3
|
Aadacl4fm5
|
AADACL4 family member 5 |
| chr2_+_119181703 | 1.01 |
ENSMUST00000028780.4
|
Chac1
|
ChaC, cation transport regulator 1 |
| chr19_+_6450641 | 0.99 |
ENSMUST00000113467.2
|
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr6_+_149031668 | 0.95 |
ENSMUST00000087348.4
|
Gm10203
|
predicted gene 10203 |
| chr3_-_129616183 | 0.95 |
ENSMUST00000196902.5
|
Rrh
|
retinal pigment epithelium derived rhodopsin homolog |
| chr8_-_21392510 | 0.94 |
ENSMUST00000122025.9
|
Gm15056
|
predicted gene 15056 |
| chr7_+_43474819 | 0.93 |
ENSMUST00000107967.3
|
Klk6
|
kallikrein related-peptidase 6 |
| chr14_+_54422156 | 0.92 |
ENSMUST00000103707.2
|
Traj34
|
T cell receptor alpha joining 34 |
| chr7_+_37883216 | 0.82 |
ENSMUST00000177983.2
|
1600014C10Rik
|
RIKEN cDNA 1600014C10 gene |
| chr9_+_19047613 | 0.82 |
ENSMUST00000215699.2
|
Olfr837
|
olfactory receptor 837 |
| chr1_-_13197387 | 0.81 |
ENSMUST00000047577.7
|
Prdm14
|
PR domain containing 14 |
| chr6_+_124908439 | 0.79 |
ENSMUST00000032214.14
|
Mlf2
|
myeloid leukemia factor 2 |
| chr9_-_76120939 | 0.78 |
ENSMUST00000074880.6
|
Gfral
|
GDNF family receptor alpha like |
| chr6_+_3993774 | 0.77 |
ENSMUST00000031673.7
|
Gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr2_-_76671491 | 0.77 |
ENSMUST00000137854.8
|
Ttn
|
titin |
| chr6_+_124908341 | 0.76 |
ENSMUST00000203021.3
|
Mlf2
|
myeloid leukemia factor 2 |
| chr7_+_17799863 | 0.73 |
ENSMUST00000108487.9
|
Ceacam12
|
carcinoembryonic antigen-related cell adhesion molecule 12 |
| chr2_-_34951443 | 0.73 |
ENSMUST00000028233.7
|
Hc
|
hemolytic complement |
| chr10_+_79590910 | 0.69 |
ENSMUST00000219981.2
ENSMUST00000219228.2 ENSMUST00000020577.4 |
Fgf22
|
fibroblast growth factor 22 |
| chr2_-_90123185 | 0.66 |
ENSMUST00000117141.4
|
Olfr1272
|
olfactory receptor 1272 |
| chr12_+_119356318 | 0.66 |
ENSMUST00000221866.2
|
Macc1
|
metastasis associated in colon cancer 1 |
| chr6_+_124908389 | 0.62 |
ENSMUST00000180095.4
|
Mlf2
|
myeloid leukemia factor 2 |
| chr7_-_127534601 | 0.60 |
ENSMUST00000141385.7
ENSMUST00000156152.3 |
Prss36
|
protease, serine 36 |
| chr7_-_103142086 | 0.54 |
ENSMUST00000055787.8
|
Olfr609
|
olfactory receptor 609 |
| chr16_-_20141815 | 0.53 |
ENSMUST00000023513.12
|
Cyp2ab1
|
cytochrome P450, family 2, subfamily ab, polypeptide 1 |
| chr17_+_25588241 | 0.52 |
ENSMUST00000160377.8
ENSMUST00000160485.8 |
Tpsg1
|
tryptase gamma 1 |
| chr12_-_114160354 | 0.51 |
ENSMUST00000103476.2
ENSMUST00000191693.2 |
Ighv3-3
|
immunoglobulin heavy variable V3-3 |
| chr11_-_87805829 | 0.50 |
ENSMUST00000074874.5
|
Olfr464
|
olfactory receptor 464 |
| chr4_-_101750986 | 0.50 |
ENSMUST00000051043.4
|
C130073F10Rik
|
RIKEN cDNA C130073F10 gene |
| chr3_+_138911648 | 0.43 |
ENSMUST00000062306.7
|
Stpg2
|
sperm tail PG rich repeat containing 2 |
| chr6_-_72357398 | 0.40 |
ENSMUST00000101285.10
ENSMUST00000074231.6 |
Vamp5
|
vesicle-associated membrane protein 5 |
| chr11_-_34048538 | 0.39 |
ENSMUST00000223852.2
|
4930469K13Rik
|
RIKEN cDNA 4930469K13 gene |
| chr2_-_88587017 | 0.38 |
ENSMUST00000099813.2
|
Olfr1199
|
olfactory receptor 1199 |
| chr6_+_132572871 | 0.37 |
ENSMUST00000076061.4
ENSMUST00000178961.2 |
Prp2
|
proline rich protein 2 |
| chr1_+_20801127 | 0.36 |
ENSMUST00000027061.5
|
Il17a
|
interleukin 17A |
| chr17_+_25115461 | 0.34 |
ENSMUST00000024978.7
|
Nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr17_+_35268942 | 0.31 |
ENSMUST00000007257.10
|
Clic1
|
chloride intracellular channel 1 |
| chr1_-_153726930 | 0.31 |
ENSMUST00000059607.8
|
Teddm2
|
transmembrane epididymal family member 2 |
| chr3_-_92493507 | 0.28 |
ENSMUST00000194965.6
|
Smcp
|
sperm mitochondria-associated cysteine-rich protein |
| chr12_-_114117264 | 0.24 |
ENSMUST00000103461.5
|
Ighv7-3
|
immunoglobulin heavy variable 7-3 |
| chr7_-_107357104 | 0.24 |
ENSMUST00000208217.2
ENSMUST00000052438.8 |
Cyb5r2
|
cytochrome b5 reductase 2 |
| chr7_+_17799849 | 0.23 |
ENSMUST00000032520.9
|
Ceacam12
|
carcinoembryonic antigen-related cell adhesion molecule 12 |
| chr19_+_29388312 | 0.23 |
ENSMUST00000112576.4
|
Pdcd1lg2
|
programmed cell death 1 ligand 2 |
| chr12_-_54703281 | 0.22 |
ENSMUST00000056228.8
|
Sptssa
|
serine palmitoyltransferase, small subunit A |
| chr2_+_87686206 | 0.21 |
ENSMUST00000217376.2
|
Olfr1151
|
olfactory receptor 1151 |
| chr9_-_106421834 | 0.19 |
ENSMUST00010181660.1
ENSMUST00000215525.2 |
ENSMUSG00001074846.1
ENSMUSG00000118396.3
|
IQ motif containing F3 IQ motif containing F3 |
| chr7_+_17799889 | 0.18 |
ENSMUST00000108483.2
|
Ceacam12
|
carcinoembryonic antigen-related cell adhesion molecule 12 |
| chr19_-_38212544 | 0.17 |
ENSMUST00000067167.6
|
Fra10ac1
|
FRA10AC1 homolog (human) |
| chr1_+_54289833 | 0.17 |
ENSMUST00000027128.11
|
Ccdc150
|
coiled-coil domain containing 150 |
| chr7_-_102591782 | 0.12 |
ENSMUST00000210571.2
ENSMUST00000064830.6 |
Olfr573-ps1
|
olfactory receptor 573, pseudogene 1 |
| chr2_-_104324035 | 0.12 |
ENSMUST00000111124.8
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr3_+_138911419 | 0.11 |
ENSMUST00000106239.8
|
Stpg2
|
sperm tail PG rich repeat containing 2 |
| chr9_+_108782646 | 0.11 |
ENSMUST00000112070.8
|
Col7a1
|
collagen, type VII, alpha 1 |
| chr7_+_27879650 | 0.06 |
ENSMUST00000172467.8
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr19_-_10952009 | 0.02 |
ENSMUST00000191343.7
|
Ms4a10
|
membrane-spanning 4-domains, subfamily A, member 10 |
| chr19_+_27194757 | 0.01 |
ENSMUST00000047645.13
ENSMUST00000167487.8 |
Vldlr
|
very low density lipoprotein receptor |
| chr3_+_145643760 | 0.01 |
ENSMUST00000039571.14
|
2410004B18Rik
|
RIKEN cDNA 2410004B18 gene |
| chr9_+_108782664 | 0.00 |
ENSMUST00000026740.6
|
Col7a1
|
collagen, type VII, alpha 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.3 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 1.5 | 4.4 | GO:0018931 | naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 0.7 | 4.4 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.6 | 5.2 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.5 | 2.2 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.4 | 4.6 | GO:0009133 | nucleoside diphosphate biosynthetic process(GO:0009133) |
| 0.3 | 1.3 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.3 | 1.3 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.3 | 1.2 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.3 | 2.1 | GO:1901748 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.3 | 1.6 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.2 | 6.9 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.2 | 0.8 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.2 | 1.5 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 1.0 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.2 | 1.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.2 | 0.8 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.1 | 0.7 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.1 | 2.0 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.1 | 1.3 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.1 | 4.1 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.1 | 3.1 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.1 | 0.9 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 2.2 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.1 | 0.4 | GO:0032747 | positive regulation of necrotic cell death(GO:0010940) positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 2.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.3 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 2.2 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.8 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 1.4 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.9 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 1.2 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 1.3 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 1.6 | GO:0007127 | meiosis I(GO:0007127) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 2.1 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 1.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 0.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 1.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 3.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 2.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 5.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 1.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 2.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.2 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.6 | 4.4 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.4 | 1.5 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.3 | 1.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.3 | 1.6 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 6.9 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.2 | 4.6 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.2 | 2.1 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.2 | 4.4 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.2 | 1.0 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.2 | 3.1 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 1.6 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.9 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.8 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 1.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 3.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 2.4 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 2.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 5.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.3 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.4 | PID IL27 PATHWAY | IL27-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 15.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 0.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 0.9 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 2.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.7 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 1.3 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 2.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |