Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Six6
Z-value: 1.23
Transcription factors associated with Six6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Six6
|
ENSMUSG00000021099.7 | Six6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Six6 | mm39_v1_chr12_+_72986665_72986674 | 0.18 | 1.3e-01 | Click! |
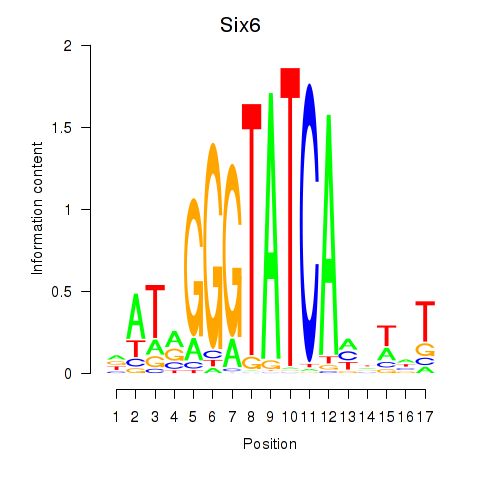
Activity profile of Six6 motif
Sorted Z-values of Six6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Six6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_70675416 | 12.47 |
ENSMUST00000103403.3
|
Igkv3-2
|
immunoglobulin kappa variable 3-2 |
| chr6_-_69792108 | 9.36 |
ENSMUST00000103367.3
|
Igkv12-44
|
immunoglobulin kappa variable 12-44 |
| chr6_-_69741999 | 9.06 |
ENSMUST00000103365.3
|
Igkv12-46
|
immunoglobulin kappa variable 12-46 |
| chr12_-_113379925 | 8.94 |
ENSMUST00000194162.6
ENSMUST00000192250.2 |
Ighd
|
immunoglobulin heavy constant delta |
| chr12_-_113386312 | 8.14 |
ENSMUST00000177715.8
ENSMUST00000103426.3 |
Ighm
|
immunoglobulin heavy constant mu |
| chr3_-_101195213 | 7.46 |
ENSMUST00000029456.5
|
Cd2
|
CD2 antigen |
| chr6_-_70313491 | 7.42 |
ENSMUST00000103388.4
|
Igkv6-20
|
immunoglobulin kappa variable 6-20 |
| chr12_-_114252202 | 7.26 |
ENSMUST00000195124.6
ENSMUST00000103481.3 |
Ighv3-6
|
immunoglobulin heavy variable 3-6 |
| chr6_+_68916540 | 7.19 |
ENSMUST00000103339.2
|
Igkv13-84
|
immunoglobulin kappa chain variable 13-84 |
| chr17_+_48554786 | 7.07 |
ENSMUST00000048065.6
|
Trem3
|
triggering receptor expressed on myeloid cells 3 |
| chr19_+_4204605 | 6.65 |
ENSMUST00000061086.9
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C polypeptide-associated protein |
| chr15_+_79779218 | 6.57 |
ENSMUST00000023054.14
|
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr7_+_130633776 | 5.82 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr11_-_102255999 | 5.73 |
ENSMUST00000006749.10
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
| chr12_-_114710326 | 5.32 |
ENSMUST00000103507.2
|
Ighv1-22
|
immunoglobulin heavy variable 1-22 |
| chr12_-_115611981 | 5.14 |
ENSMUST00000103540.3
ENSMUST00000199266.2 |
Ighv8-12
|
immunoglobulin heavy variable V8-12 |
| chr17_+_41121979 | 5.04 |
ENSMUST00000024721.8
ENSMUST00000233740.2 |
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr4_+_11758147 | 5.02 |
ENSMUST00000029871.12
ENSMUST00000108303.2 |
Cdh17
|
cadherin 17 |
| chr3_-_113166153 | 5.00 |
ENSMUST00000098673.5
|
Amy2a5
|
amylase 2a5 |
| chr3_+_87283687 | 4.93 |
ENSMUST00000163661.8
ENSMUST00000072480.9 |
Fcrl1
|
Fc receptor-like 1 |
| chr12_-_114646685 | 4.72 |
ENSMUST00000194350.6
ENSMUST00000103504.3 |
Ighv1-18
|
immunoglobulin heavy variable V1-18 |
| chr3_+_90173813 | 4.68 |
ENSMUST00000098914.10
|
Dennd4b
|
DENN/MADD domain containing 4B |
| chr1_-_165535654 | 4.65 |
ENSMUST00000097474.9
|
Rcsd1
|
RCSD domain containing 1 |
| chr6_+_67816777 | 4.57 |
ENSMUST00000200578.5
ENSMUST00000103308.3 |
Igkv9-129
|
immunoglobulin kappa variable 9-129 |
| chr3_+_87283767 | 4.48 |
ENSMUST00000194786.6
ENSMUST00000191666.2 |
Fcrl1
|
Fc receptor-like 1 |
| chr1_+_60785517 | 4.47 |
ENSMUST00000027165.3
|
Cd28
|
CD28 antigen |
| chr2_+_127178072 | 4.36 |
ENSMUST00000028846.7
|
Dusp2
|
dual specificity phosphatase 2 |
| chr11_+_117688486 | 4.34 |
ENSMUST00000106331.2
|
6030468B19Rik
|
RIKEN cDNA 6030468B19 gene |
| chr1_+_139382485 | 4.25 |
ENSMUST00000200083.5
ENSMUST00000053364.12 |
Aspm
|
abnormal spindle microtubule assembly |
| chr3_+_87283748 | 4.24 |
ENSMUST00000167200.7
|
Fcrl1
|
Fc receptor-like 1 |
| chr15_+_101152078 | 4.20 |
ENSMUST00000228985.2
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr15_-_74635423 | 4.19 |
ENSMUST00000040404.8
|
Ly6d
|
lymphocyte antigen 6 complex, locus D |
| chr12_-_113589576 | 4.12 |
ENSMUST00000103446.2
|
Ighv5-6
|
immunoglobulin heavy variable 5-6 |
| chr3_-_113263974 | 3.82 |
ENSMUST00000098667.5
|
Amy2a2
|
amylase 2a2 |
| chr7_+_43086432 | 3.77 |
ENSMUST00000070518.4
|
Nkg7
|
natural killer cell group 7 sequence |
| chr14_+_53607470 | 3.72 |
ENSMUST00000103652.5
|
Trav14n-3
|
T cell receptor alpha variable 14N-3 |
| chr7_+_43086554 | 3.71 |
ENSMUST00000206741.2
|
Nkg7
|
natural killer cell group 7 sequence |
| chr3_-_75177378 | 3.58 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr4_+_156098292 | 3.54 |
ENSMUST00000030952.6
|
Tnfrsf4
|
tumor necrosis factor receptor superfamily, member 4 |
| chr14_+_53791444 | 3.49 |
ENSMUST00000198297.2
|
Trav14-1
|
T cell receptor alpha variable 14-1 |
| chr6_-_68887957 | 3.47 |
ENSMUST00000200454.2
|
Igkv4-86
|
immunoglobulin kappa variable 4-86 |
| chr7_-_119058489 | 3.26 |
ENSMUST00000207887.3
ENSMUST00000239424.2 ENSMUST00000033255.8 |
Gp2
|
glycoprotein 2 (zymogen granule membrane) |
| chrX_-_141749704 | 3.23 |
ENSMUST00000041317.3
|
Ammecr1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr17_+_34524884 | 3.20 |
ENSMUST00000074557.11
|
H2-Eb1
|
histocompatibility 2, class II antigen E beta |
| chr4_-_134099840 | 3.15 |
ENSMUST00000030643.3
|
Extl1
|
exostosin-like glycosyltransferase 1 |
| chr7_-_100504610 | 2.94 |
ENSMUST00000156855.8
|
Relt
|
RELT tumor necrosis factor receptor |
| chr8_+_22224506 | 2.89 |
ENSMUST00000080533.6
|
Defa24
|
defensin, alpha, 24 |
| chr18_+_89224219 | 2.87 |
ENSMUST00000236835.2
|
Cd226
|
CD226 antigen |
| chr11_-_6180127 | 2.81 |
ENSMUST00000004505.3
|
Npc1l1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr14_+_53093071 | 2.71 |
ENSMUST00000181038.3
ENSMUST00000187138.2 |
Trav14d-1
|
T cell receptor alpha variable 14D-1 |
| chr14_+_53628092 | 2.70 |
ENSMUST00000103636.4
|
Trav7-2
|
T cell receptor alpha variable 7-2 |
| chr2_+_59314989 | 2.68 |
ENSMUST00000028369.6
|
Dapl1
|
death associated protein-like 1 |
| chr11_+_49327451 | 2.68 |
ENSMUST00000215226.2
|
Olfr1388
|
olfactory receptor 1388 |
| chr1_-_173703424 | 2.67 |
ENSMUST00000186442.7
|
Mndal
|
myeloid nuclear differentiation antigen like |
| chr1_-_154692678 | 2.65 |
ENSMUST00000238369.2
|
Cacna1e
|
calcium channel, voltage-dependent, R type, alpha 1E subunit |
| chr4_+_102843540 | 2.54 |
ENSMUST00000030248.12
ENSMUST00000125417.9 ENSMUST00000169211.3 |
Dynlt5
|
dynein light chain Tctex-type 5 |
| chr12_-_103409912 | 2.54 |
ENSMUST00000055071.9
|
Ifi27l2a
|
interferon, alpha-inducible protein 27 like 2A |
| chr6_+_123099615 | 2.50 |
ENSMUST00000161636.8
ENSMUST00000161365.8 |
Clec4a2
|
C-type lectin domain family 4, member a2 |
| chr8_+_70275079 | 2.49 |
ENSMUST00000164890.8
ENSMUST00000034325.6 ENSMUST00000238452.2 |
Lpar2
|
lysophosphatidic acid receptor 2 |
| chr3_-_37778470 | 2.49 |
ENSMUST00000108105.2
ENSMUST00000079755.5 ENSMUST00000099128.2 |
Gm5148
|
predicted gene 5148 |
| chr2_+_131333672 | 2.47 |
ENSMUST00000028806.12
|
Smox
|
spermine oxidase |
| chr17_+_36176485 | 2.40 |
ENSMUST00000127442.8
ENSMUST00000144382.8 |
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr9_+_32547529 | 2.40 |
ENSMUST00000238819.2
|
Ets1
|
E26 avian leukemia oncogene 1, 5' domain |
| chr7_+_127661835 | 2.36 |
ENSMUST00000106242.10
ENSMUST00000120355.8 ENSMUST00000106240.9 ENSMUST00000098015.10 |
Itgam
Gm49368
|
integrin alpha M predicted gene, 49368 |
| chr14_-_66071412 | 2.30 |
ENSMUST00000022613.10
|
Esco2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr9_+_106099797 | 2.30 |
ENSMUST00000062241.11
|
Tlr9
|
toll-like receptor 9 |
| chr7_-_44198157 | 2.25 |
ENSMUST00000145956.2
ENSMUST00000049343.15 |
Pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr5_-_23821523 | 2.25 |
ENSMUST00000088392.9
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr8_+_57908920 | 2.21 |
ENSMUST00000034023.4
|
Scrg1
|
scrapie responsive gene 1 |
| chr5_-_108943211 | 2.20 |
ENSMUST00000004943.2
|
Tmed11
|
transmembrane p24 trafficking protein 11 |
| chr12_-_114451189 | 2.18 |
ENSMUST00000103493.3
|
Ighv1-4
|
immunoglobulin heavy variable 1-4 |
| chr14_+_52921421 | 2.15 |
ENSMUST00000200127.2
|
Trav7d-2
|
T cell receptor alpha variable 7D-2 |
| chr6_-_120893771 | 2.13 |
ENSMUST00000004560.12
|
Bid
|
BH3 interacting domain death agonist |
| chr8_+_70285133 | 2.12 |
ENSMUST00000081503.13
|
Pbx4
|
pre B cell leukemia homeobox 4 |
| chr6_+_123100382 | 2.11 |
ENSMUST00000032248.8
|
Clec4a2
|
C-type lectin domain family 4, member a2 |
| chr14_+_53878158 | 2.10 |
ENSMUST00000179267.4
|
Trav14-2
|
T cell receptor alpha variable 14-2 |
| chr9_+_108820846 | 2.10 |
ENSMUST00000198140.5
ENSMUST00000051873.15 |
Pfkfb4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr1_+_60948307 | 2.05 |
ENSMUST00000097720.4
|
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr6_+_34389269 | 2.04 |
ENSMUST00000007449.9
|
Akr1b7
|
aldo-keto reductase family 1, member B7 |
| chr8_+_70285282 | 2.01 |
ENSMUST00000131637.9
|
Pbx4
|
pre B cell leukemia homeobox 4 |
| chr5_+_122239830 | 1.96 |
ENSMUST00000146733.5
|
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr7_+_89814713 | 1.96 |
ENSMUST00000207084.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr6_+_123100272 | 1.92 |
ENSMUST00000041779.13
|
Clec4a2
|
C-type lectin domain family 4, member a2 |
| chr16_+_10652910 | 1.91 |
ENSMUST00000037913.9
|
Rmi2
|
RecQ mediated genome instability 2 |
| chr15_-_82128888 | 1.89 |
ENSMUST00000089155.6
ENSMUST00000089157.11 |
Cenpm
|
centromere protein M |
| chr10_+_128073900 | 1.69 |
ENSMUST00000105245.3
|
Timeless
|
timeless circadian clock 1 |
| chr14_+_52875909 | 1.69 |
ENSMUST00000103571.2
|
Trav6-1
|
T cell receptor alpha variable 6-1 |
| chr9_+_110856425 | 1.69 |
ENSMUST00000199313.2
|
Ltf
|
lactotransferrin |
| chr17_+_35460722 | 1.66 |
ENSMUST00000068056.12
ENSMUST00000174757.8 ENSMUST00000173731.8 |
Ddx39b
|
DEAD box helicase 39b |
| chr6_+_40763875 | 1.60 |
ENSMUST00000195870.3
|
Mgam2-ps
|
maltase-glucoamylase 2, pseudogene |
| chr4_-_87724512 | 1.59 |
ENSMUST00000148059.2
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr11_+_115455260 | 1.58 |
ENSMUST00000021085.11
|
Nup85
|
nucleoporin 85 |
| chr14_+_54440591 | 1.57 |
ENSMUST00000103725.2
|
Traj16
|
T cell receptor alpha joining 16 |
| chr6_-_146855880 | 1.51 |
ENSMUST00000111622.2
ENSMUST00000036592.15 |
1700034J05Rik
|
RIKEN cDNA 1700034J05 gene |
| chr5_-_107074110 | 1.45 |
ENSMUST00000117588.8
|
Hfm1
|
HFM1, ATP-dependent DNA helicase homolog |
| chr15_-_66985760 | 1.45 |
ENSMUST00000092640.6
|
St3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr17_+_43879496 | 1.45 |
ENSMUST00000169694.2
|
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr9_-_119170456 | 1.43 |
ENSMUST00000139870.2
|
Myd88
|
myeloid differentiation primary response gene 88 |
| chr8_+_11890474 | 1.42 |
ENSMUST00000033909.14
ENSMUST00000209692.2 |
Tex29
|
testis expressed 29 |
| chr11_+_51895018 | 1.37 |
ENSMUST00000109079.9
ENSMUST00000063303.11 ENSMUST00000109077.9 ENSMUST00000109080.9 ENSMUST00000109081.9 ENSMUST00000121591.8 ENSMUST00000109078.8 ENSMUST00000063321.13 ENSMUST00000135076.8 ENSMUST00000120374.8 |
Cdkl3
|
cyclin-dependent kinase-like 3 |
| chrX_-_19347558 | 1.37 |
ENSMUST00000089246.6
|
Tex13c3
|
TEX13 family member C3 |
| chr14_+_53157900 | 1.35 |
ENSMUST00000178252.3
|
Trav9d-3
|
T cell receptor alpha variable 9D-3 |
| chr6_+_41369290 | 1.33 |
ENSMUST00000049079.9
|
Gm5771
|
predicted gene 5771 |
| chrX_-_100777806 | 1.32 |
ENSMUST00000056614.7
|
Cxcr3
|
chemokine (C-X-C motif) receptor 3 |
| chr7_-_133304244 | 1.31 |
ENSMUST00000209636.2
ENSMUST00000153698.3 |
Uros
|
uroporphyrinogen III synthase |
| chr19_+_7589890 | 1.28 |
ENSMUST00000025929.12
ENSMUST00000239394.2 ENSMUST00000148558.3 |
Plaat5
|
phospholipase A and acyltransferase 5 |
| chr7_+_43625387 | 1.27 |
ENSMUST00000081399.4
|
Klk1b9
|
kallikrein 1-related peptidase b9 |
| chr8_-_4325886 | 1.27 |
ENSMUST00000003029.14
|
Timm44
|
translocase of inner mitochondrial membrane 44 |
| chr11_-_106163753 | 1.26 |
ENSMUST00000021052.16
|
Smarcd2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 |
| chr11_-_103099349 | 1.26 |
ENSMUST00000174567.3
ENSMUST00000021323.11 ENSMUST00000107026.9 |
Efcab15
|
EF-hand calcium binding domain 15 |
| chr17_+_43879366 | 1.25 |
ENSMUST00000167418.8
|
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr11_+_57994964 | 1.25 |
ENSMUST00000020822.12
|
Cnot8
|
CCR4-NOT transcription complex, subunit 8 |
| chr5_-_123126550 | 1.23 |
ENSMUST00000086200.11
ENSMUST00000156474.8 |
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr14_+_54082691 | 1.20 |
ENSMUST00000103674.6
|
Trav19
|
T cell receptor alpha variable 19 |
| chr15_-_82872073 | 1.20 |
ENSMUST00000229439.2
|
Tcf20
|
transcription factor 20 |
| chr1_+_152683568 | 1.19 |
ENSMUST00000190323.7
|
Ncf2
|
neutrophil cytosolic factor 2 |
| chr13_+_100261358 | 1.19 |
ENSMUST00000022147.15
ENSMUST00000091321.6 ENSMUST00000143937.2 |
Smn1
|
survival motor neuron 1 |
| chr7_-_101552989 | 1.19 |
ENSMUST00000106969.9
|
Tomt
|
transmembrane O-methyltransferase |
| chr16_-_65359406 | 1.16 |
ENSMUST00000231259.2
|
Chmp2b
|
charged multivesicular body protein 2B |
| chr13_+_19362068 | 1.14 |
ENSMUST00000103553.3
|
Trgv7
|
T cell receptor gamma, variable 7 |
| chr1_-_131066004 | 1.14 |
ENSMUST00000016670.9
|
Dyrk3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chr7_-_140480314 | 1.09 |
ENSMUST00000026561.10
|
Cox8b
|
cytochrome c oxidase subunit 8B |
| chr7_-_97827461 | 1.06 |
ENSMUST00000040971.14
|
Capn5
|
calpain 5 |
| chr7_-_104991477 | 1.05 |
ENSMUST00000213290.2
|
Olfr691
|
olfactory receptor 691 |
| chr15_-_73702827 | 1.04 |
ENSMUST00000238402.2
|
Mroh5
|
maestro heat-like repeat family member 5 |
| chr2_-_114031897 | 1.04 |
ENSMUST00000050668.4
|
Zfp770
|
zinc finger protein 770 |
| chr2_+_117080212 | 1.03 |
ENSMUST00000028825.5
|
Fam98b
|
family with sequence similarity 98, member B |
| chrX_-_33580888 | 1.02 |
ENSMUST00000238632.2
|
Btbd35f9
|
BTB domain containing 35, family member 9 |
| chr2_-_89855921 | 1.01 |
ENSMUST00000216616.3
|
Olfr1264
|
olfactory receptor 1264 |
| chr17_+_47221377 | 1.01 |
ENSMUST00000024773.6
|
Prph2
|
peripherin 2 |
| chr8_-_94262434 | 1.00 |
ENSMUST00000212722.2
|
Ces5a
|
carboxylesterase 5A |
| chr14_-_75830550 | 0.99 |
ENSMUST00000164082.9
ENSMUST00000169658.9 |
Cby2
|
chibby family member 2 |
| chr11_-_99121822 | 0.98 |
ENSMUST00000103133.4
|
Smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr1_+_165312738 | 0.96 |
ENSMUST00000111440.8
ENSMUST00000027852.15 ENSMUST00000111439.8 |
Adcy10
|
adenylate cyclase 10 |
| chr3_+_94305824 | 0.95 |
ENSMUST00000050975.6
|
Lingo4
|
leucine rich repeat and Ig domain containing 4 |
| chr4_+_119052548 | 0.94 |
ENSMUST00000106345.3
|
Svbp
|
small vasohibin binding protein |
| chr14_+_53878403 | 0.94 |
ENSMUST00000184874.2
|
Trav14-2
|
T cell receptor alpha variable 14-2 |
| chrX_+_101971975 | 0.90 |
ENSMUST00000211116.2
|
1700018G05Rik
|
RIKEN cDNA 1700018G05 gene |
| chr14_+_53315909 | 0.90 |
ENSMUST00000103608.4
|
Trav14d-3-dv8
|
T cell receptor alpha variable 14D-3-DV8 |
| chr1_-_59134042 | 0.90 |
ENSMUST00000238601.2
ENSMUST00000238949.2 ENSMUST00000097080.4 |
C2cd6
|
C2 calcium dependent domain containing 6 |
| chr13_-_56283331 | 0.89 |
ENSMUST00000045788.9
ENSMUST00000016081.13 |
Macroh2a1
|
macroH2A.1 histone |
| chr19_-_11313471 | 0.87 |
ENSMUST00000056035.9
ENSMUST00000067532.11 |
Ms4a7
|
membrane-spanning 4-domains, subfamily A, member 7 |
| chr7_+_80764547 | 0.87 |
ENSMUST00000026820.11
|
Slc28a1
|
solute carrier family 28 (sodium-coupled nucleoside transporter), member 1 |
| chr17_-_53846438 | 0.87 |
ENSMUST00000056198.4
|
Pp2d1
|
protein phosphatase 2C-like domain containing 1 |
| chr13_-_23806530 | 0.86 |
ENSMUST00000062045.4
|
H1f4
|
H1.4 linker histone, cluster member |
| chr19_-_6002210 | 0.85 |
ENSMUST00000236013.2
|
Pola2
|
polymerase (DNA directed), alpha 2 |
| chr6_-_69920632 | 0.85 |
ENSMUST00000198880.5
ENSMUST00000103371.3 |
Igkv12-38
|
immunoglobulin kappa chain variable 12-38 |
| chr5_-_109207435 | 0.83 |
ENSMUST00000164875.3
|
Vmn2r11
|
vomeronasal 2, receptor 11 |
| chr4_-_58499398 | 0.82 |
ENSMUST00000107570.2
|
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr3_-_96634880 | 0.80 |
ENSMUST00000029741.9
|
Polr3c
|
polymerase (RNA) III (DNA directed) polypeptide C |
| chrX_-_12539778 | 0.80 |
ENSMUST00000060108.7
|
1810030O07Rik
|
RIKEN cDNA 1810030O07 gene |
| chr5_+_77163869 | 0.80 |
ENSMUST00000031161.11
ENSMUST00000117880.8 |
Thegl
|
theg spermatid protein like |
| chr3_-_9069745 | 0.79 |
ENSMUST00000120143.8
|
Tpd52
|
tumor protein D52 |
| chr14_-_70864666 | 0.78 |
ENSMUST00000022694.17
|
Dmtn
|
dematin actin binding protein |
| chr6_+_38639945 | 0.78 |
ENSMUST00000114874.5
|
Clec2l
|
C-type lectin domain family 2, member L |
| chr5_+_88032867 | 0.77 |
ENSMUST00000129757.9
|
Odam
|
odontogenic, ameloblast asssociated |
| chr4_-_97472844 | 0.75 |
ENSMUST00000107067.8
ENSMUST00000107068.9 |
E130114P18Rik
|
RIKEN cDNA E130114P18 gene |
| chr7_-_79974166 | 0.75 |
ENSMUST00000047362.11
ENSMUST00000121882.8 |
Rccd1
|
RCC1 domain containing 1 |
| chr18_-_43820759 | 0.75 |
ENSMUST00000082254.8
|
Jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr7_-_103799746 | 0.73 |
ENSMUST00000059121.5
|
Ubqlnl
|
ubiquilin-like |
| chr2_-_89408791 | 0.73 |
ENSMUST00000217402.2
|
Olfr1245
|
olfactory receptor 1245 |
| chr8_-_23143422 | 0.73 |
ENSMUST00000033938.7
|
Polb
|
polymerase (DNA directed), beta |
| chr9_+_77543776 | 0.72 |
ENSMUST00000057781.8
|
Klhl31
|
kelch-like 31 |
| chr10_+_73657689 | 0.72 |
ENSMUST00000064562.14
ENSMUST00000193174.6 ENSMUST00000105426.10 ENSMUST00000129404.9 ENSMUST00000131321.9 ENSMUST00000126920.9 ENSMUST00000147189.9 ENSMUST00000105424.10 ENSMUST00000092420.13 ENSMUST00000105429.10 ENSMUST00000193361.6 ENSMUST00000131724.9 ENSMUST00000152655.9 ENSMUST00000151116.9 ENSMUST00000155701.9 ENSMUST00000152819.9 ENSMUST00000125517.9 ENSMUST00000124046.8 ENSMUST00000146682.8 ENSMUST00000177107.8 ENSMUST00000149977.9 ENSMUST00000191854.6 |
Pcdh15
|
protocadherin 15 |
| chrX_+_31608531 | 0.71 |
ENSMUST00000238528.2
|
Btbd35f22
|
BTB domain containing 35, family member 22 |
| chr3_+_96634974 | 0.70 |
ENSMUST00000029740.14
|
Rnf115
|
ring finger protein 115 |
| chr16_+_29028860 | 0.70 |
ENSMUST00000162747.8
|
Plaat1
|
phospholipase A and acyltransferase 1 |
| chr6_-_120893725 | 0.69 |
ENSMUST00000145948.2
|
Bid
|
BH3 interacting domain death agonist |
| chr8_+_3443018 | 0.68 |
ENSMUST00000004684.13
ENSMUST00000145394.3 |
Arhgef18
|
rho/rac guanine nucleotide exchange factor (GEF) 18 |
| chr19_+_9824919 | 0.67 |
ENSMUST00000179814.3
|
Scgb2a2
|
secretoglobin, family 2A, member 2 |
| chr2_+_37078210 | 0.66 |
ENSMUST00000213969.2
|
Olfr365
|
olfactory receptor 365 |
| chr7_-_24423715 | 0.66 |
ENSMUST00000081657.6
|
Lypd11
|
Ly6/PLAUR domain containing 11 |
| chrX_-_104973003 | 0.66 |
ENSMUST00000130980.2
ENSMUST00000113573.8 |
Atrx
|
ATRX, chromatin remodeler |
| chrX_-_153673263 | 0.65 |
ENSMUST00000096852.5
|
Cldn34b1
|
claudin 34B1 |
| chr18_+_32948436 | 0.65 |
ENSMUST00000025237.5
|
Tslp
|
thymic stromal lymphopoietin |
| chr16_-_65359566 | 0.65 |
ENSMUST00000004965.8
|
Chmp2b
|
charged multivesicular body protein 2B |
| chr1_+_97697881 | 0.64 |
ENSMUST00000112844.10
ENSMUST00000112842.8 ENSMUST00000027571.13 |
Gin1
|
gypsy retrotransposon integrase 1 |
| chrX_-_56438322 | 0.63 |
ENSMUST00000114730.8
|
Rbmx
|
RNA binding motif protein, X chromosome |
| chr18_+_56565188 | 0.63 |
ENSMUST00000070166.6
|
Gramd3
|
GRAM domain containing 3 |
| chr2_-_180284468 | 0.60 |
ENSMUST00000037877.11
|
Tcfl5
|
transcription factor-like 5 (basic helix-loop-helix) |
| chr1_-_128287347 | 0.60 |
ENSMUST00000190495.2
ENSMUST00000027601.11 |
Mcm6
|
minichromosome maintenance complex component 6 |
| chr3_-_104684798 | 0.60 |
ENSMUST00000139783.2
|
Tafa3
|
TAFA chemokine like family member 3 |
| chr1_-_39844467 | 0.59 |
ENSMUST00000171319.4
|
Gm3646
|
predicted gene 3646 |
| chr3_+_32760447 | 0.59 |
ENSMUST00000194781.6
|
Actl6a
|
actin-like 6A |
| chr9_+_120400510 | 0.58 |
ENSMUST00000165532.3
|
Rpl14
|
ribosomal protein L14 |
| chrX_+_32070863 | 0.58 |
ENSMUST00000238237.2
|
Btbd35f1
|
BTB domain containing 35, family member 1 |
| chr1_+_134313676 | 0.57 |
ENSMUST00000162187.2
|
Mgat4f
|
MGAT4 family, member F |
| chrX_+_32411401 | 0.57 |
ENSMUST00000178747.3
|
Btbd35f5
|
BTB domain containing 35, family member 5 |
| chrX_+_32750842 | 0.56 |
ENSMUST00000178827.3
|
Btbd35f12
|
BTB domain containing 35, family member 12 |
| chrX_-_33394003 | 0.55 |
ENSMUST00000179466.2
|
Btbd35f6
|
BTB domain containing 35, family member 6 |
| chrX_-_33014777 | 0.54 |
ENSMUST00000186329.2
|
Btbd35f15
|
BTB domain containing 35, family member 15 |
| chr17_-_14279793 | 0.54 |
ENSMUST00000186636.3
|
Gm7358
|
predicted gene 7358 |
| chr18_-_9282754 | 0.53 |
ENSMUST00000041007.4
|
Gjd4
|
gap junction protein, delta 4 |
| chrX_-_33139812 | 0.52 |
ENSMUST00000105117.3
|
Btbd35f14
|
BTB domain containing 35, family member 14 |
| chr8_-_85740220 | 0.52 |
ENSMUST00000209322.2
|
Best2
|
bestrophin 2 |
| chr11_+_100210705 | 0.52 |
ENSMUST00000049385.14
|
Eif1
|
eukaryotic translation initiation factor 1 |
| chr4_-_75196485 | 0.51 |
ENSMUST00000030103.9
|
Dmac1
|
distal membrane arm assembly complex 1 |
| chr8_+_21382681 | 0.51 |
ENSMUST00000098908.4
|
Defb33
|
defensin beta 33 |
| chr6_+_56947258 | 0.50 |
ENSMUST00000226130.2
ENSMUST00000228276.2 |
Vmn1r5
|
vomeronasal 1 receptor 5 |
| chr7_+_108265625 | 0.50 |
ENSMUST00000213979.3
ENSMUST00000216331.2 ENSMUST00000217170.2 |
Olfr510
|
olfactory receptor 510 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.1 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 1.7 | 5.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 1.3 | 5.0 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 1.1 | 3.3 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.9 | 7.1 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.8 | 4.7 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.8 | 2.3 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.8 | 2.3 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.7 | 4.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.7 | 2.0 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.6 | 9.7 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.5 | 2.7 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.5 | 4.2 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.5 | 4.7 | GO:0071474 | cellular hyperosmotic response(GO:0071474) |
| 0.5 | 7.5 | GO:2000484 | positive regulation of interferon-gamma secretion(GO:1902715) positive regulation of interleukin-8 secretion(GO:2000484) |
| 0.5 | 2.9 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.5 | 2.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.4 | 1.3 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.4 | 1.7 | GO:0044407 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.4 | 2.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.4 | 1.4 | GO:2000338 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.4 | 3.5 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.3 | 4.5 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.3 | 2.0 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.3 | 1.7 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.3 | 2.4 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.3 | 2.0 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.3 | 2.8 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.3 | 6.6 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.3 | 1.2 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.3 | 2.7 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.3 | 2.5 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.3 | 2.4 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.2 | 1.7 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.2 | 1.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.2 | 0.5 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.2 | 5.8 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.2 | 0.9 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.2 | 2.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 0.8 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.2 | 5.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 2.8 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.2 | 1.2 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.2 | 28.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.2 | 0.7 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.2 | 3.2 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.2 | 4.2 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.1 | 1.2 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.1 | 0.8 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.1 | 0.6 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 2.2 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 1.3 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) |
| 0.1 | 1.6 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 1.3 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 1.8 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 2.5 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 1.5 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 21.0 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 0.5 | GO:0014717 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.1 | 4.4 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.1 | 0.2 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 0.4 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.1 | 1.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 1.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.3 | GO:0035128 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.1 | 0.3 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.0 | 0.7 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.6 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.5 | GO:0009048 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 2.7 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.3 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.1 | GO:0098942 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 0.0 | 0.2 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 1.6 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.4 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 1.4 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.9 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.2 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.2 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) negative regulation of monocyte differentiation(GO:0045656) |
| 0.0 | 1.6 | GO:0033045 | regulation of sister chromatid segregation(GO:0033045) |
| 0.0 | 0.5 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.4 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.9 | GO:0010596 | negative regulation of endothelial cell migration(GO:0010596) |
| 0.0 | 0.9 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.0 | 3.1 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.2 | GO:0003351 | epithelial cilium movement(GO:0003351) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 17.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.6 | 2.3 | GO:0036019 | endolysosome(GO:0036019) |
| 0.5 | 2.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.4 | 5.8 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.4 | 4.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.3 | 1.7 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.3 | 2.0 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.3 | 3.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 28.7 | GO:0019814 | immunoglobulin complex(GO:0019814) immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 8.9 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.2 | 6.4 | GO:0001741 | XY body(GO:0001741) |
| 0.2 | 1.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 1.7 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 0.9 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 2.8 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 1.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 1.6 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.8 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 2.7 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 7.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.6 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 2.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 1.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 1.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.6 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 1.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 7.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 5.7 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 2.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.6 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.8 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 1.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 8.5 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 2.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 2.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 2.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 1.4 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.0 | 1.2 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 1.5 | GO:0000776 | kinetochore(GO:0000776) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0035375 | zymogen binding(GO:0035375) |
| 1.3 | 8.8 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 1.3 | 5.0 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 1.1 | 6.6 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 1.0 | 3.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.8 | 2.5 | GO:0052895 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 0.8 | 7.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.4 | 1.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.4 | 1.6 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.4 | 1.4 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.3 | 45.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.3 | 2.4 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.3 | 1.7 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.3 | 0.9 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.3 | 2.7 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.3 | 1.6 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.3 | 5.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.3 | 4.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 2.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.2 | 1.3 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.2 | 5.0 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.2 | 2.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 6.5 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 1.3 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.2 | 1.4 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.2 | 0.5 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 15.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 2.0 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 2.7 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 2.3 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.1 | 4.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.8 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 2.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 2.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.6 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 1.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.4 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 4.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.2 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.1 | 1.7 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 1.0 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 1.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 4.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 2.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.3 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.1 | 0.4 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 2.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.4 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 1.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 2.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.8 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 2.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 1.1 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 1.2 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 1.5 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 1.1 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.6 | GO:0008026 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 0.2 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 3.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 2.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 2.6 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 4.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 2.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 4.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 2.4 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 3.5 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 2.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 1.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.3 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.6 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.8 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.2 | 4.5 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.2 | 2.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 2.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 5.0 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 2.7 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 0.9 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.1 | 2.0 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.1 | 4.2 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 2.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 2.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 9.8 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 1.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 0.7 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 2.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.8 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.5 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 2.8 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 5.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.8 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 2.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 2.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.7 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.4 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |