Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
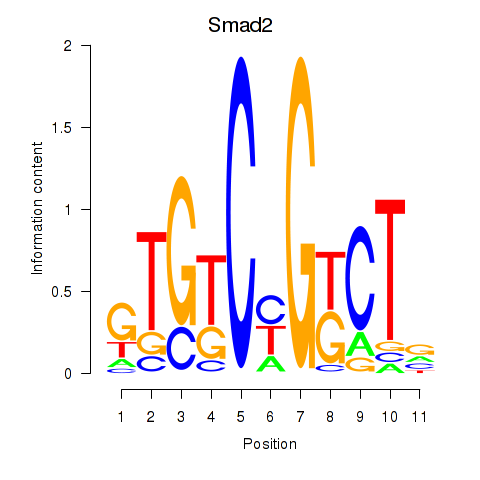
Results for Smad2
Z-value: 0.66
Transcription factors associated with Smad2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Smad2
|
ENSMUSG00000024563.17 | Smad2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Smad2 | mm39_v1_chr18_+_76375206_76375245 | 0.05 | 6.5e-01 | Click! |
Activity profile of Smad2 motif
Sorted Z-values of Smad2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Smad2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_126062272 | 7.57 |
ENSMUST00000032974.13
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr7_+_19144950 | 7.28 |
ENSMUST00000208710.2
ENSMUST00000003643.3 |
Ckm
|
creatine kinase, muscle |
| chr1_-_172125555 | 4.50 |
ENSMUST00000085913.11
ENSMUST00000097464.4 |
Atp1a2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr1_-_173195236 | 3.45 |
ENSMUST00000005470.5
ENSMUST00000111220.8 |
Cadm3
|
cell adhesion molecule 3 |
| chr12_-_103304573 | 2.99 |
ENSMUST00000149431.2
|
Asb2
|
ankyrin repeat and SOCS box-containing 2 |
| chr4_+_42950367 | 2.91 |
ENSMUST00000084662.12
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr16_-_34334314 | 2.62 |
ENSMUST00000151491.8
ENSMUST00000114960.9 |
Kalrn
|
kalirin, RhoGEF kinase |
| chr3_-_116762617 | 2.46 |
ENSMUST00000143611.2
ENSMUST00000040097.14 |
Palmd
|
palmdelphin |
| chr6_+_30639217 | 2.44 |
ENSMUST00000031806.10
|
Cpa1
|
carboxypeptidase A1, pancreatic |
| chr7_+_98916635 | 2.40 |
ENSMUST00000122101.8
ENSMUST00000068973.11 ENSMUST00000207883.2 |
Map6
|
microtubule-associated protein 6 |
| chr15_+_82136598 | 2.35 |
ENSMUST00000136948.3
|
1500009C09Rik
|
RIKEN cDNA 1500009C09 gene |
| chr12_+_51640097 | 2.31 |
ENSMUST00000164782.10
ENSMUST00000085412.7 |
Coch
|
cochlin |
| chr15_-_101833160 | 2.26 |
ENSMUST00000023797.8
|
Krt4
|
keratin 4 |
| chr16_-_34334454 | 2.24 |
ENSMUST00000089655.12
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr10_-_5019044 | 2.21 |
ENSMUST00000095899.5
|
Syne1
|
spectrin repeat containing, nuclear envelope 1 |
| chr11_+_63022328 | 2.19 |
ENSMUST00000018361.10
|
Pmp22
|
peripheral myelin protein 22 |
| chr17_-_46343291 | 2.01 |
ENSMUST00000071648.12
ENSMUST00000142351.9 |
Vegfa
|
vascular endothelial growth factor A |
| chr5_-_113938417 | 1.84 |
ENSMUST00000160374.2
ENSMUST00000067853.6 |
Tmem119
|
transmembrane protein 119 |
| chr4_+_117706390 | 1.74 |
ENSMUST00000132043.9
ENSMUST00000169990.8 |
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr7_+_98916677 | 1.63 |
ENSMUST00000127492.2
|
Map6
|
microtubule-associated protein 6 |
| chr13_-_78347876 | 1.57 |
ENSMUST00000091458.13
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr8_-_69636825 | 1.55 |
ENSMUST00000185176.8
|
Lzts1
|
leucine zipper, putative tumor suppressor 1 |
| chr3_+_87878378 | 1.55 |
ENSMUST00000090973.12
|
Nes
|
nestin |
| chr3_+_87878436 | 1.48 |
ENSMUST00000160694.2
|
Nes
|
nestin |
| chr4_-_155095441 | 1.42 |
ENSMUST00000105631.9
ENSMUST00000139976.9 ENSMUST00000145662.9 |
Plch2
|
phospholipase C, eta 2 |
| chr9_+_110074574 | 1.42 |
ENSMUST00000197850.5
|
Cspg5
|
chondroitin sulfate proteoglycan 5 |
| chr5_-_146521629 | 1.37 |
ENSMUST00000200112.2
ENSMUST00000197431.2 ENSMUST00000197825.2 |
Gpr12
|
G-protein coupled receptor 12 |
| chr18_+_37847792 | 1.36 |
ENSMUST00000192511.2
ENSMUST00000193476.2 |
Pcdhga7
|
protocadherin gamma subfamily A, 7 |
| chr11_-_33097400 | 1.35 |
ENSMUST00000020507.8
|
Fgf18
|
fibroblast growth factor 18 |
| chr4_+_117706559 | 1.28 |
ENSMUST00000163288.2
|
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr11_-_96720738 | 1.28 |
ENSMUST00000107657.8
ENSMUST00000081775.12 |
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1 |
| chr3_-_116762476 | 1.25 |
ENSMUST00000119557.8
|
Palmd
|
palmdelphin |
| chr7_+_142030744 | 1.21 |
ENSMUST00000149521.8
|
Lsp1
|
lymphocyte specific 1 |
| chr11_+_69737491 | 1.17 |
ENSMUST00000019605.4
|
Plscr3
|
phospholipid scramblase 3 |
| chr7_+_28151370 | 1.15 |
ENSMUST00000190954.7
|
Lrfn1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr10_-_49664839 | 1.08 |
ENSMUST00000220263.2
ENSMUST00000218823.2 |
Grik2
|
glutamate receptor, ionotropic, kainate 2 (beta 2) |
| chr6_+_117818135 | 1.04 |
ENSMUST00000112859.8
ENSMUST00000137224.8 ENSMUST00000164472.8 ENSMUST00000112861.8 ENSMUST00000223041.2 |
Zfp637
|
zinc finger protein 637 |
| chr6_-_6882068 | 1.03 |
ENSMUST00000142635.2
ENSMUST00000052609.9 |
Dlx5
|
distal-less homeobox 5 |
| chrX_+_98864627 | 1.01 |
ENSMUST00000096363.3
|
Tmem28
|
transmembrane protein 28 |
| chr12_+_55883101 | 1.00 |
ENSMUST00000059250.8
|
Brms1l
|
breast cancer metastasis-suppressor 1-like |
| chr6_+_117818216 | 0.98 |
ENSMUST00000136889.8
ENSMUST00000134526.8 ENSMUST00000112858.8 ENSMUST00000112860.2 |
Zfp637
|
zinc finger protein 637 |
| chr8_+_66838927 | 0.96 |
ENSMUST00000039540.12
ENSMUST00000110253.3 |
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr1_+_75456173 | 0.96 |
ENSMUST00000113575.9
ENSMUST00000148980.2 ENSMUST00000050899.7 ENSMUST00000187411.2 |
Tmem198
|
transmembrane protein 198 |
| chr11_+_69737437 | 0.95 |
ENSMUST00000152566.8
ENSMUST00000108633.9 |
Plscr3
|
phospholipid scramblase 3 |
| chr13_-_104953631 | 0.94 |
ENSMUST00000154165.2
|
Cwc27
|
CWC27 spliceosome-associated protein |
| chrX_-_20797736 | 0.93 |
ENSMUST00000001156.8
|
Cfp
|
complement factor properdin |
| chr2_-_148285450 | 0.84 |
ENSMUST00000099269.4
|
Cd93
|
CD93 antigen |
| chr11_+_101010764 | 0.81 |
ENSMUST00000043680.9
|
Tubg1
|
tubulin, gamma 1 |
| chr15_-_84007373 | 0.77 |
ENSMUST00000230566.2
ENSMUST00000019012.4 |
Pnpla5
|
patatin-like phospholipase domain containing 5 |
| chr13_-_100240570 | 0.77 |
ENSMUST00000038104.12
|
Bdp1
|
B double prime 1, subunit of RNA polymerase III transcription initiation factor IIIB |
| chr1_-_150868802 | 0.76 |
ENSMUST00000074783.12
ENSMUST00000137197.9 |
Hmcn1
|
hemicentin 1 |
| chr4_+_117692583 | 0.75 |
ENSMUST00000169885.8
|
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr14_+_10123804 | 0.72 |
ENSMUST00000022262.6
ENSMUST00000224714.2 |
Fezf2
|
Fez family zinc finger 2 |
| chr2_+_173918715 | 0.71 |
ENSMUST00000087908.10
ENSMUST00000044638.13 ENSMUST00000156054.2 |
Stx16
|
syntaxin 16 |
| chr9_+_35332837 | 0.71 |
ENSMUST00000119129.10
|
Cdon
|
cell adhesion molecule-related/down-regulated by oncogenes |
| chr13_-_104953370 | 0.68 |
ENSMUST00000022228.13
|
Cwc27
|
CWC27 spliceosome-associated protein |
| chr6_-_90758954 | 0.64 |
ENSMUST00000238821.2
|
Iqsec1
|
IQ motif and Sec7 domain 1 |
| chr14_+_4137837 | 0.50 |
ENSMUST00000022296.7
|
Ube2e1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr17_-_24752683 | 0.50 |
ENSMUST00000061764.14
|
Rab26
|
RAB26, member RAS oncogene family |
| chr15_-_101471324 | 0.49 |
ENSMUST00000023714.5
|
Krt90
|
keratin 90 |
| chr14_+_4137944 | 0.48 |
ENSMUST00000225612.2
|
Ube2e1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr4_+_83335947 | 0.48 |
ENSMUST00000030206.10
ENSMUST00000071544.11 |
Snapc3
|
small nuclear RNA activating complex, polypeptide 3 |
| chr6_-_124733441 | 0.41 |
ENSMUST00000088357.12
|
Atn1
|
atrophin 1 |
| chr18_+_37440497 | 0.40 |
ENSMUST00000056712.4
|
Pcdhb4
|
protocadherin beta 4 |
| chr8_+_19343376 | 0.39 |
ENSMUST00000033852.8
|
Defb3
|
defensin beta 3 |
| chr17_+_3447465 | 0.39 |
ENSMUST00000072156.7
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr3_+_58484057 | 0.38 |
ENSMUST00000107924.3
|
Selenot
|
selenoprotein T |
| chr5_+_115769960 | 0.36 |
ENSMUST00000031492.15
|
Rab35
|
RAB35, member RAS oncogene family |
| chr10_-_37014859 | 0.35 |
ENSMUST00000092584.6
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr11_-_101010715 | 0.31 |
ENSMUST00000017946.6
|
Retreg3
|
reticulophagy regulator family member 3 |
| chr6_+_83114020 | 0.30 |
ENSMUST00000121093.8
|
Rtkn
|
rhotekin |
| chr12_-_115129697 | 0.30 |
ENSMUST00000103524.2
ENSMUST00000193145.2 |
Ighv8-6
|
immunoglobulin heavy variable V8-6 |
| chr6_+_83114086 | 0.29 |
ENSMUST00000087938.11
|
Rtkn
|
rhotekin |
| chr6_-_142647985 | 0.27 |
ENSMUST00000205202.3
ENSMUST00000073173.12 ENSMUST00000111771.8 |
Abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr12_-_114370581 | 0.25 |
ENSMUST00000191918.2
|
Ighv6-4
|
immunoglobulin heavy variable V6-4 |
| chr7_-_99276310 | 0.24 |
ENSMUST00000178124.3
|
Tpbgl
|
trophoblast glycoprotein-like |
| chr1_-_160079007 | 0.24 |
ENSMUST00000191909.6
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr12_-_113294138 | 0.24 |
ENSMUST00000194304.6
ENSMUST00000103420.3 |
Ighg1
|
immunoglobulin heavy constant gamma 1 (G1m marker) |
| chr7_-_44702269 | 0.20 |
ENSMUST00000057293.8
|
Prr12
|
proline rich 12 |
| chr6_-_55152002 | 0.20 |
ENSMUST00000003569.6
|
Inmt
|
indolethylamine N-methyltransferase |
| chr16_-_18165876 | 0.12 |
ENSMUST00000125287.9
|
Tango2
|
transport and golgi organization 2 |
| chr16_+_18166045 | 0.07 |
ENSMUST00000239533.1
ENSMUST00000239534.1 |
ARVCF
|
armadillo repeat deleted in velocardiofacial syndrome |
| chr6_-_142647944 | 0.07 |
ENSMUST00000100827.5
ENSMUST00000087527.11 |
Abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr17_-_34834745 | 0.03 |
ENSMUST00000168353.4
|
Egfl8
|
EGF-like domain 8 |
| chr1_+_153767478 | 0.03 |
ENSMUST00000050660.6
|
Teddm1a
|
transmembrane epididymal protein 1A |
| chr12_-_113271532 | 0.02 |
ENSMUST00000192188.3
ENSMUST00000103418.3 |
Ighg2b
|
immunoglobulin heavy constant gamma 2B |
| chr13_+_104953679 | 0.02 |
ENSMUST00000022230.15
|
Srek1ip1
|
splicing regulatory glutamine/lysine-rich protein 1interacting protein 1 |
| chr3_+_135144202 | 0.01 |
ENSMUST00000166033.6
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr11_-_75081284 | 0.01 |
ENSMUST00000044949.11
|
Dph1
|
diphthamide biosynthesis 1 |
| chr11_-_101010640 | 0.00 |
ENSMUST00000107295.10
|
Retreg3
|
reticulophagy regulator family member 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.6 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
| 1.3 | 3.8 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 1.1 | 4.5 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.7 | 2.0 | GO:0038190 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 0.6 | 4.9 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.3 | 1.0 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.3 | 2.2 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.2 | 1.3 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.2 | 1.8 | GO:1903012 | positive regulation of bone development(GO:1903012) |
| 0.1 | 0.8 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 1.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.7 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 7.3 | GO:0042398 | cellular modified amino acid biosynthetic process(GO:0042398) |
| 0.1 | 0.2 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.1 | 0.7 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 2.2 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 1.0 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 1.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 1.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.4 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.4 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.5 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.0 | 0.7 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.8 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 1.5 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 4.0 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 3.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 3.0 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 2.1 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.4 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 5.4 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.8 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.0 | GO:0001796 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type I hypersensitivity(GO:0001812) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.0 | 0.6 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 2.9 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 2.3 | GO:0050680 | negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.0 | 1.0 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 2.8 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.0 | 0.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 7.6 | GO:0031673 | H zone(GO:0031673) |
| 0.4 | 4.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.4 | 3.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 2.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 1.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 0.8 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 2.2 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 0.8 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.1 | 0.4 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 2.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.0 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 3.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.4 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 2.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 5.3 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 4.9 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.4 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 4.3 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 0.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.6 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 3.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 7.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.7 | 2.0 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.6 | 7.6 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.4 | 4.5 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) steroid hormone binding(GO:1990239) |
| 0.3 | 1.0 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.3 | 1.3 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.3 | 3.8 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.3 | 0.8 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.3 | 3.0 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.2 | 1.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 0.5 | GO:0019002 | GMP binding(GO:0019002) |
| 0.2 | 1.0 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 4.9 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.8 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 1.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 2.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 2.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 2.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 1.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 1.4 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 1.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 2.9 | GO:0051087 | unfolded protein binding(GO:0051082) chaperone binding(GO:0051087) |
| 0.0 | 0.4 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 1.0 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 1.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 5.6 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 4.9 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 7.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 2.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 2.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.2 | 2.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 3.8 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 4.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 1.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 0.8 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 5.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.0 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 2.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 7.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |