Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Smad4
Z-value: 1.09
Transcription factors associated with Smad4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Smad4
|
ENSMUSG00000024515.14 | Smad4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Smad4 | mm39_v1_chr18_-_73836810_73836877 | 0.09 | 4.3e-01 | Click! |
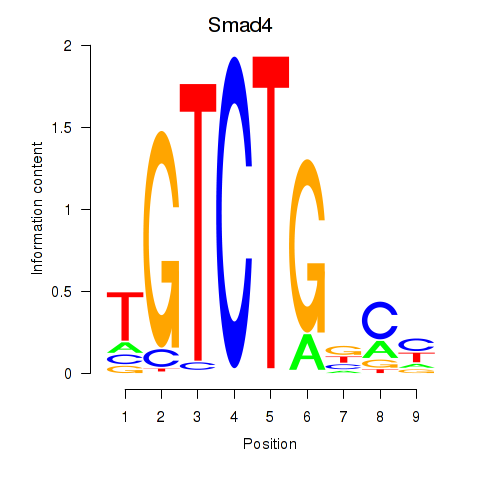
Activity profile of Smad4 motif
Sorted Z-values of Smad4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Smad4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_45847344 | 21.36 |
ENSMUST00000034590.4
|
Tagln
|
transgelin |
| chr6_+_146934082 | 6.54 |
ENSMUST00000036194.6
|
Rep15
|
RAB15 effector protein |
| chr16_+_34605282 | 6.36 |
ENSMUST00000023538.9
|
Mylk
|
myosin, light polypeptide kinase |
| chr17_+_34482183 | 6.09 |
ENSMUST00000040828.7
ENSMUST00000237342.2 ENSMUST00000237866.2 |
H2-Ab1
|
histocompatibility 2, class II antigen A, beta 1 |
| chrX_+_55824797 | 5.81 |
ENSMUST00000114773.10
|
Fhl1
|
four and a half LIM domains 1 |
| chr11_+_105866030 | 5.52 |
ENSMUST00000001964.8
|
Ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| chrX_-_7537580 | 5.51 |
ENSMUST00000033486.6
|
Plp2
|
proteolipid protein 2 |
| chr1_-_74130488 | 5.39 |
ENSMUST00000187584.7
|
Tns1
|
tensin 1 |
| chr7_-_70010341 | 5.37 |
ENSMUST00000032768.15
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr6_-_52217821 | 5.37 |
ENSMUST00000121043.2
|
Hoxa10
|
homeobox A10 |
| chr10_+_18720760 | 5.35 |
ENSMUST00000019998.9
|
Perp
|
PERP, TP53 apoptosis effector |
| chr12_-_103304573 | 4.87 |
ENSMUST00000149431.2
|
Asb2
|
ankyrin repeat and SOCS box-containing 2 |
| chr17_+_75485791 | 4.79 |
ENSMUST00000135447.8
ENSMUST00000112516.8 |
Ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr6_-_68681962 | 4.64 |
ENSMUST00000103330.2
|
Igkv10-94
|
immunoglobulin kappa variable 10-94 |
| chr19_-_40260060 | 4.56 |
ENSMUST00000068439.13
|
Pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr18_-_52662917 | 4.38 |
ENSMUST00000171470.8
|
Lox
|
lysyl oxidase |
| chr18_-_52662728 | 4.37 |
ENSMUST00000025409.9
|
Lox
|
lysyl oxidase |
| chr7_+_130633776 | 4.21 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr10_+_79986280 | 4.21 |
ENSMUST00000153477.8
|
Midn
|
midnolin |
| chr4_+_138694422 | 4.16 |
ENSMUST00000116094.5
ENSMUST00000239443.2 |
Rnf186
|
ring finger protein 186 |
| chr2_+_125876566 | 4.15 |
ENSMUST00000064794.14
|
Fgf7
|
fibroblast growth factor 7 |
| chr17_+_75485906 | 4.15 |
ENSMUST00000112514.2
|
Ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr2_+_69500444 | 4.09 |
ENSMUST00000100050.4
|
Klhl41
|
kelch-like 41 |
| chr19_-_40260286 | 4.09 |
ENSMUST00000182432.2
|
Pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr19_+_5790918 | 4.03 |
ENSMUST00000081496.6
|
Ltbp3
|
latent transforming growth factor beta binding protein 3 |
| chr11_+_67131403 | 3.90 |
ENSMUST00000170942.2
|
Myh4
|
myosin, heavy polypeptide 4, skeletal muscle |
| chr12_-_112766266 | 3.83 |
ENSMUST00000239525.1
|
Ahnak2
|
AHNAK nucleoprotein 2 |
| chr5_+_92831468 | 3.83 |
ENSMUST00000168878.8
|
Shroom3
|
shroom family member 3 |
| chr14_-_25903524 | 3.81 |
ENSMUST00000052286.16
|
Plac9a
|
placenta specific 9a |
| chr2_+_152578164 | 3.73 |
ENSMUST00000038368.9
ENSMUST00000109824.2 |
Id1
|
inhibitor of DNA binding 1, HLH protein |
| chr5_+_64960705 | 3.70 |
ENSMUST00000165536.8
|
Klf3
|
Kruppel-like factor 3 (basic) |
| chr5_+_31070739 | 3.69 |
ENSMUST00000031055.8
|
Emilin1
|
elastin microfibril interfacer 1 |
| chr6_+_113448388 | 3.45 |
ENSMUST00000058300.14
|
Il17rc
|
interleukin 17 receptor C |
| chr2_+_125876883 | 3.38 |
ENSMUST00000110442.2
|
Fgf7
|
fibroblast growth factor 7 |
| chr19_-_23425757 | 3.37 |
ENSMUST00000036069.8
|
Mamdc2
|
MAM domain containing 2 |
| chr15_+_58381886 | 3.35 |
ENSMUST00000161028.2
|
Fer1l6
|
fer-1-like 6 (C. elegans) |
| chr7_+_18817767 | 3.34 |
ENSMUST00000032568.14
ENSMUST00000122999.8 ENSMUST00000108473.10 ENSMUST00000108474.2 ENSMUST00000238982.2 |
Dmpk
|
dystrophia myotonica-protein kinase |
| chr2_-_163592127 | 3.26 |
ENSMUST00000017841.4
|
Ada
|
adenosine deaminase |
| chr6_+_113449237 | 3.23 |
ENSMUST00000204447.3
|
Il17rc
|
interleukin 17 receptor C |
| chr7_+_45063079 | 3.19 |
ENSMUST00000058879.8
|
Ntf5
|
neurotrophin 5 |
| chr17_+_34372046 | 3.18 |
ENSMUST00000114232.4
|
H2-DMb1
|
histocompatibility 2, class II, locus Mb1 |
| chr4_-_148584906 | 3.15 |
ENSMUST00000030840.4
|
Angptl7
|
angiopoietin-like 7 |
| chr19_-_6947112 | 3.13 |
ENSMUST00000025912.10
|
Plcb3
|
phospholipase C, beta 3 |
| chr19_-_6947076 | 3.07 |
ENSMUST00000237074.2
|
Plcb3
|
phospholipase C, beta 3 |
| chr11_-_101062111 | 2.95 |
ENSMUST00000164474.8
ENSMUST00000043397.14 |
Plekhh3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr4_+_150171822 | 2.94 |
ENSMUST00000094451.4
|
Gpr157
|
G protein-coupled receptor 157 |
| chr2_+_27567246 | 2.93 |
ENSMUST00000166775.8
|
Rxra
|
retinoid X receptor alpha |
| chr1_+_171509565 | 2.85 |
ENSMUST00000015499.14
ENSMUST00000068584.7 |
Cd48
|
CD48 antigen |
| chr4_-_94940425 | 2.77 |
ENSMUST00000107094.2
|
Jun
|
jun proto-oncogene |
| chr11_-_33097400 | 2.73 |
ENSMUST00000020507.8
|
Fgf18
|
fibroblast growth factor 18 |
| chr2_-_164231015 | 2.73 |
ENSMUST00000167427.2
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr7_+_44865177 | 2.69 |
ENSMUST00000033060.14
ENSMUST00000210447.2 ENSMUST00000211744.2 ENSMUST00000107801.10 |
Tead2
|
TEA domain family member 2 |
| chr12_-_114286421 | 2.68 |
ENSMUST00000103483.3
|
Ighv3-8
|
immunoglobulin heavy variable V3-8 |
| chr3_+_89344006 | 2.67 |
ENSMUST00000038942.10
ENSMUST00000130858.8 ENSMUST00000146630.8 ENSMUST00000145753.2 |
Pbxip1
|
pre B cell leukemia transcription factor interacting protein 1 |
| chr6_-_69800923 | 2.66 |
ENSMUST00000103368.3
|
Igkv5-43
|
immunoglobulin kappa chain variable 5-43 |
| chr2_-_155676765 | 2.57 |
ENSMUST00000029143.7
ENSMUST00000239423.2 |
Fam83c
|
family with sequence similarity 83, member C |
| chr10_+_39488930 | 2.48 |
ENSMUST00000019987.7
|
Traf3ip2
|
TRAF3 interacting protein 2 |
| chrX_+_21581135 | 2.47 |
ENSMUST00000033414.8
|
Slc6a14
|
solute carrier family 6 (neurotransmitter transporter), member 14 |
| chr4_-_133981387 | 2.44 |
ENSMUST00000060050.6
|
Grrp1
|
glycine/arginine rich protein 1 |
| chr15_-_103275190 | 2.44 |
ENSMUST00000023128.8
|
Itga5
|
integrin alpha 5 (fibronectin receptor alpha) |
| chr13_-_22016364 | 2.38 |
ENSMUST00000102979.2
|
H4c18
|
H4 clustered histone 18 |
| chr12_-_36206780 | 2.33 |
ENSMUST00000223382.2
ENSMUST00000020856.6 |
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr12_-_36206750 | 2.30 |
ENSMUST00000221388.2
|
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr15_+_102885467 | 2.30 |
ENSMUST00000001706.7
|
Hoxc9
|
homeobox C9 |
| chr14_-_54651442 | 2.24 |
ENSMUST00000227334.2
|
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr17_-_46342739 | 2.24 |
ENSMUST00000024747.14
|
Vegfa
|
vascular endothelial growth factor A |
| chr2_+_27567213 | 2.20 |
ENSMUST00000077257.12
|
Rxra
|
retinoid X receptor alpha |
| chr7_+_25386418 | 2.19 |
ENSMUST00000002678.10
|
Tgfb1
|
transforming growth factor, beta 1 |
| chr7_+_140796537 | 2.15 |
ENSMUST00000141804.2
|
Rassf7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr6_-_112364974 | 2.14 |
ENSMUST00000238755.2
ENSMUST00000060847.6 |
Ssu2
|
ssu-2 homolog (C. elegans) |
| chr10_-_37014859 | 2.11 |
ENSMUST00000092584.6
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chrX_-_71318353 | 2.11 |
ENSMUST00000064780.4
|
Gabre
|
gamma-aminobutyric acid (GABA) A receptor, subunit epsilon |
| chr6_+_68414401 | 2.08 |
ENSMUST00000103324.3
|
Igkv15-103
|
immunoglobulin kappa chain variable 15-103 |
| chr4_+_8691303 | 2.07 |
ENSMUST00000051558.10
|
Chd7
|
chromodomain helicase DNA binding protein 7 |
| chr5_-_138980690 | 2.04 |
ENSMUST00000046901.14
ENSMUST00000076095.14 |
Pdgfa
|
platelet derived growth factor, alpha |
| chr9_-_58065800 | 2.01 |
ENSMUST00000168864.4
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr17_+_34816826 | 2.01 |
ENSMUST00000015596.10
ENSMUST00000174496.9 ENSMUST00000173992.8 |
Ager
|
advanced glycosylation end product-specific receptor |
| chr19_+_4282487 | 2.00 |
ENSMUST00000235306.2
|
Pold4
|
polymerase (DNA-directed), delta 4 |
| chr17_-_71158703 | 1.98 |
ENSMUST00000166395.9
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr5_-_41921834 | 1.95 |
ENSMUST00000060820.8
|
Nkx3-2
|
NK3 homeobox 2 |
| chr8_+_96078886 | 1.95 |
ENSMUST00000034243.7
|
Mmp15
|
matrix metallopeptidase 15 |
| chr1_-_153061758 | 1.93 |
ENSMUST00000185356.7
|
Lamc2
|
laminin, gamma 2 |
| chr7_-_81642034 | 1.92 |
ENSMUST00000026096.10
|
Bnc1
|
basonuclin 1 |
| chr15_+_102884874 | 1.88 |
ENSMUST00000173306.2
|
Hoxc9
|
homeobox C9 |
| chr9_+_108782664 | 1.84 |
ENSMUST00000026740.6
|
Col7a1
|
collagen, type VII, alpha 1 |
| chr12_-_36206626 | 1.84 |
ENSMUST00000220828.2
|
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr15_-_96358612 | 1.84 |
ENSMUST00000047835.8
|
Scaf11
|
SR-related CTD-associated factor 11 |
| chr1_+_171197660 | 1.81 |
ENSMUST00000111286.9
|
Nectin4
|
nectin cell adhesion molecule 4 |
| chr9_+_7344357 | 1.81 |
ENSMUST00000150167.2
ENSMUST00000120655.8 |
Mmp12
|
matrix metallopeptidase 12 |
| chr7_+_101043568 | 1.81 |
ENSMUST00000098243.4
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr6_+_38410848 | 1.80 |
ENSMUST00000160583.8
|
Ubn2
|
ubinuclein 2 |
| chr10_-_118705029 | 1.78 |
ENSMUST00000004281.10
|
Dyrk2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr3_-_135397298 | 1.77 |
ENSMUST00000029812.14
|
Nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 1, p105 |
| chr10_+_90918802 | 1.75 |
ENSMUST00000020150.10
ENSMUST00000020149.6 |
Ikbip
|
IKBKB interacting protein |
| chr7_+_45055077 | 1.75 |
ENSMUST00000107774.3
|
Kcna7
|
potassium voltage-gated channel, shaker-related subfamily, member 7 |
| chr19_+_4282040 | 1.74 |
ENSMUST00000237518.2
|
Pold4
|
polymerase (DNA-directed), delta 4 |
| chr9_+_108782646 | 1.74 |
ENSMUST00000112070.8
|
Col7a1
|
collagen, type VII, alpha 1 |
| chr11_-_3881995 | 1.72 |
ENSMUST00000020710.11
ENSMUST00000109989.10 ENSMUST00000109991.8 ENSMUST00000109993.9 |
Tcn2
|
transcobalamin 2 |
| chr17_-_25300112 | 1.72 |
ENSMUST00000024984.7
|
Tmem204
|
transmembrane protein 204 |
| chr3_+_60436570 | 1.71 |
ENSMUST00000192607.6
|
Mbnl1
|
muscleblind like splicing factor 1 |
| chrX_+_98179725 | 1.71 |
ENSMUST00000052839.7
|
Efnb1
|
ephrin B1 |
| chr2_-_173117936 | 1.70 |
ENSMUST00000139306.2
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr6_-_129428746 | 1.69 |
ENSMUST00000204012.2
ENSMUST00000037481.10 |
Clec1a
|
C-type lectin domain family 1, member a |
| chr19_-_31641770 | 1.68 |
ENSMUST00000073581.6
|
Prkg1
|
protein kinase, cGMP-dependent, type I |
| chr13_+_21995906 | 1.68 |
ENSMUST00000104941.4
|
H4c17
|
H4 clustered histone 17 |
| chr12_-_113928438 | 1.66 |
ENSMUST00000103478.4
|
Ighv3-1
|
immunoglobulin heavy variable 3-1 |
| chr9_+_61280501 | 1.66 |
ENSMUST00000162583.8
ENSMUST00000161993.8 ENSMUST00000160882.8 ENSMUST00000160724.8 ENSMUST00000162973.8 ENSMUST00000159050.8 |
Tle3
|
transducin-like enhancer of split 3 |
| chr14_-_55950545 | 1.65 |
ENSMUST00000002389.9
ENSMUST00000226907.2 |
Tgm1
|
transglutaminase 1, K polypeptide |
| chr5_+_88523967 | 1.65 |
ENSMUST00000073363.2
|
Amtn
|
amelotin |
| chr1_+_187730018 | 1.64 |
ENSMUST00000027906.13
|
Esrrg
|
estrogen-related receptor gamma |
| chr9_+_53678801 | 1.63 |
ENSMUST00000048670.10
|
Slc35f2
|
solute carrier family 35, member F2 |
| chr11_-_3881789 | 1.62 |
ENSMUST00000109992.8
ENSMUST00000109988.2 |
Tcn2
|
transcobalamin 2 |
| chr11_-_69304501 | 1.54 |
ENSMUST00000094077.5
|
Kdm6b
|
KDM1 lysine (K)-specific demethylase 6B |
| chr13_+_23715220 | 1.52 |
ENSMUST00000102972.6
|
H4c8
|
H4 clustered histone 8 |
| chr6_+_15720653 | 1.51 |
ENSMUST00000101663.10
ENSMUST00000190255.7 ENSMUST00000189359.7 ENSMUST00000125326.8 |
Mdfic
|
MyoD family inhibitor domain containing |
| chr15_+_102012782 | 1.50 |
ENSMUST00000230474.2
|
Tns2
|
tensin 2 |
| chr12_-_11200306 | 1.46 |
ENSMUST00000055673.2
|
Kcns3
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
| chr11_-_3881960 | 1.46 |
ENSMUST00000109990.8
|
Tcn2
|
transcobalamin 2 |
| chr17_-_46343291 | 1.42 |
ENSMUST00000071648.12
ENSMUST00000142351.9 |
Vegfa
|
vascular endothelial growth factor A |
| chr15_-_98505508 | 1.42 |
ENSMUST00000096224.6
|
Adcy6
|
adenylate cyclase 6 |
| chr19_+_4281953 | 1.39 |
ENSMUST00000025773.5
|
Pold4
|
polymerase (DNA-directed), delta 4 |
| chr19_+_4147391 | 1.39 |
ENSMUST00000174514.2
ENSMUST00000174149.8 |
Cdk2ap2
|
CDK2-associated protein 2 |
| chr1_+_53100796 | 1.38 |
ENSMUST00000027269.7
ENSMUST00000191197.2 |
Mstn
|
myostatin |
| chr9_+_61280764 | 1.35 |
ENSMUST00000160541.8
ENSMUST00000161207.8 ENSMUST00000159630.8 |
Tle3
|
transducin-like enhancer of split 3 |
| chr3_+_63203516 | 1.35 |
ENSMUST00000029400.7
|
Mme
|
membrane metallo endopeptidase |
| chr14_-_30890544 | 1.32 |
ENSMUST00000036618.14
|
Stab1
|
stabilin 1 |
| chrX_-_94070277 | 1.30 |
ENSMUST00000096367.5
|
Spin4
|
spindlin family, member 4 |
| chr8_-_41469786 | 1.26 |
ENSMUST00000117735.8
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr4_-_133972890 | 1.22 |
ENSMUST00000030644.8
|
Zfp593
|
zinc finger protein 593 |
| chr4_-_148215135 | 1.21 |
ENSMUST00000030862.5
|
Draxin
|
dorsal inhibitory axon guidance protein |
| chr19_+_4015900 | 1.20 |
ENSMUST00000025794.7
|
Aldh3b3
|
aldehyde dehydrogenase 3 family, member B3 |
| chr6_-_129428869 | 1.20 |
ENSMUST00000203162.3
|
Clec1a
|
C-type lectin domain family 1, member a |
| chr15_-_98676007 | 1.18 |
ENSMUST00000226655.2
ENSMUST00000228546.2 ENSMUST00000023732.12 ENSMUST00000226610.2 |
Wnt10b
|
wingless-type MMTV integration site family, member 10B |
| chr14_+_76652369 | 1.16 |
ENSMUST00000110888.8
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr7_+_19197192 | 1.12 |
ENSMUST00000137613.9
|
Exoc3l2
|
exocyst complex component 3-like 2 |
| chr15_+_102011415 | 1.12 |
ENSMUST00000046144.10
|
Tns2
|
tensin 2 |
| chr7_-_119494918 | 1.09 |
ENSMUST00000059851.14
|
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr4_-_117035922 | 1.09 |
ENSMUST00000153953.2
ENSMUST00000106436.8 |
Kif2c
|
kinesin family member 2C |
| chr15_+_102011352 | 1.08 |
ENSMUST00000169627.9
|
Tns2
|
tensin 2 |
| chr18_-_33597060 | 1.08 |
ENSMUST00000168890.2
|
Nrep
|
neuronal regeneration related protein |
| chr11_+_69737437 | 1.07 |
ENSMUST00000152566.8
ENSMUST00000108633.9 |
Plscr3
|
phospholipid scramblase 3 |
| chr1_-_180524587 | 1.07 |
ENSMUST00000027778.8
|
Mixl1
|
Mix1 homeobox-like 1 (Xenopus laevis) |
| chr15_-_103161237 | 1.04 |
ENSMUST00000154510.8
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr14_+_65187485 | 1.03 |
ENSMUST00000043914.8
ENSMUST00000239450.2 |
Ints9
|
integrator complex subunit 9 |
| chr2_+_29759495 | 1.03 |
ENSMUST00000047521.7
ENSMUST00000134152.2 |
Cercam
|
cerebral endothelial cell adhesion molecule |
| chr10_+_79986988 | 1.03 |
ENSMUST00000146516.8
ENSMUST00000144526.2 |
Midn
|
midnolin |
| chr7_+_44866095 | 1.02 |
ENSMUST00000209437.2
|
Tead2
|
TEA domain family member 2 |
| chr1_+_59802543 | 1.00 |
ENSMUST00000087435.7
|
Bmpr2
|
bone morphogenetic protein receptor, type II (serine/threonine kinase) |
| chr5_+_76632178 | 1.00 |
ENSMUST00000172369.2
|
Gm7271
|
predicted gene 7271 |
| chr10_-_107321938 | 0.99 |
ENSMUST00000000445.2
|
Myf5
|
myogenic factor 5 |
| chr5_-_123822338 | 0.99 |
ENSMUST00000111561.8
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr8_+_79754980 | 0.98 |
ENSMUST00000087927.11
ENSMUST00000098614.9 |
Zfp827
|
zinc finger protein 827 |
| chr1_+_187730032 | 0.98 |
ENSMUST00000110938.2
|
Esrrg
|
estrogen-related receptor gamma |
| chr9_-_14292453 | 0.95 |
ENSMUST00000167549.2
|
Endod1
|
endonuclease domain containing 1 |
| chr18_+_69478790 | 0.93 |
ENSMUST00000202116.4
ENSMUST00000114982.8 ENSMUST00000078486.13 ENSMUST00000202772.4 ENSMUST00000201288.4 |
Tcf4
|
transcription factor 4 |
| chr11_+_97554192 | 0.92 |
ENSMUST00000044730.12
|
Mllt6
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 6 |
| chr3_+_102641822 | 0.90 |
ENSMUST00000029451.12
|
Tspan2
|
tetraspanin 2 |
| chr3_+_116761916 | 0.86 |
ENSMUST00000117592.2
|
4930455H04Rik
|
RIKEN cDNA 4930455H04 gene |
| chr1_-_87029312 | 0.82 |
ENSMUST00000113270.3
|
Alpi
|
alkaline phosphatase, intestinal |
| chr3_+_63203235 | 0.80 |
ENSMUST00000194134.6
|
Mme
|
membrane metallo endopeptidase |
| chr11_-_118233326 | 0.80 |
ENSMUST00000103024.4
|
Cep295nl
|
CEP295 N-terminal like |
| chr5_+_76631887 | 0.79 |
ENSMUST00000191515.8
|
Gm7271
|
predicted gene 7271 |
| chr9_+_21336300 | 0.79 |
ENSMUST00000172482.8
ENSMUST00000174050.8 |
Dnm2
|
dynamin 2 |
| chr4_-_137309415 | 0.78 |
ENSMUST00000238941.2
|
Ldlrad2
|
low density lipoprotein receptor class A domain containing 2 |
| chr14_+_54409257 | 0.77 |
ENSMUST00000197949.2
|
Traj47
|
T cell receptor alpha joining 47 |
| chr14_+_32321824 | 0.77 |
ENSMUST00000068938.7
ENSMUST00000228878.2 |
Prrxl1
|
paired related homeobox protein-like 1 |
| chr18_+_67338437 | 0.75 |
ENSMUST00000210564.3
|
Chmp1b
|
charged multivesicular body protein 1B |
| chr10_-_90918515 | 0.74 |
ENSMUST00000159110.8
|
Apaf1
|
apoptotic peptidase activating factor 1 |
| chrX_+_158491589 | 0.73 |
ENSMUST00000080394.13
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr7_+_140796559 | 0.72 |
ENSMUST00000148975.3
|
Rassf7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr11_-_55498559 | 0.70 |
ENSMUST00000108853.8
ENSMUST00000075603.5 |
Glra1
|
glycine receptor, alpha 1 subunit |
| chr13_-_98773862 | 0.69 |
ENSMUST00000050389.5
|
Tmem174
|
transmembrane protein 174 |
| chr9_+_61280890 | 0.68 |
ENSMUST00000161689.8
|
Tle3
|
transducin-like enhancer of split 3 |
| chr11_-_115310743 | 0.68 |
ENSMUST00000106537.8
ENSMUST00000043931.9 ENSMUST00000073791.10 |
Atp5h
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit D |
| chr14_-_54791816 | 0.67 |
ENSMUST00000022784.9
|
Haus4
|
HAUS augmin-like complex, subunit 4 |
| chr14_-_58063585 | 0.62 |
ENSMUST00000022536.3
|
Ska3
|
spindle and kinetochore associated complex subunit 3 |
| chr7_+_99412714 | 0.61 |
ENSMUST00000220001.2
ENSMUST00000219663.2 |
Olfr521
|
olfactory receptor 521 |
| chr13_+_111391544 | 0.60 |
ENSMUST00000054716.4
|
Actbl2
|
actin, beta-like 2 |
| chr11_-_52104814 | 0.59 |
ENSMUST00000075844.4
|
Olfr1371
|
olfactory receptor 1371 |
| chr4_-_119047146 | 0.59 |
ENSMUST00000124626.9
|
Ermap
|
erythroblast membrane-associated protein |
| chr5_-_124490296 | 0.59 |
ENSMUST00000111472.6
|
Cdk2ap1
|
CDK2 (cyclin-dependent kinase 2)-associated protein 1 |
| chr1_+_152275575 | 0.59 |
ENSMUST00000044311.9
|
Colgalt2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr11_+_69737491 | 0.58 |
ENSMUST00000019605.4
|
Plscr3
|
phospholipid scramblase 3 |
| chr7_+_100970435 | 0.57 |
ENSMUST00000210192.2
ENSMUST00000172630.8 |
Stard10
|
START domain containing 10 |
| chr5_-_123822351 | 0.57 |
ENSMUST00000111564.8
ENSMUST00000063905.12 |
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr19_-_10716335 | 0.56 |
ENSMUST00000025571.9
ENSMUST00000238167.2 |
Cd5
|
CD5 antigen |
| chr4_-_119047180 | 0.55 |
ENSMUST00000150864.3
ENSMUST00000141227.9 |
Ermap
|
erythroblast membrane-associated protein |
| chr9_+_40098375 | 0.54 |
ENSMUST00000062229.6
|
Olfr986
|
olfactory receptor 986 |
| chr13_-_23735822 | 0.52 |
ENSMUST00000102971.2
|
H4c6
|
H4 clustered histone 6 |
| chr1_-_133728779 | 0.51 |
ENSMUST00000143567.8
|
Atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr18_-_84104507 | 0.51 |
ENSMUST00000060303.10
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr3_+_127426783 | 0.50 |
ENSMUST00000029587.9
|
Neurog2
|
neurogenin 2 |
| chr12_+_36207113 | 0.49 |
ENSMUST00000041640.5
|
Ankmy2
|
ankyrin repeat and MYND domain containing 2 |
| chr17_+_28259749 | 0.49 |
ENSMUST00000233869.2
|
Anks1
|
ankyrin repeat and SAM domain containing 1 |
| chr3_-_96833336 | 0.49 |
ENSMUST00000062944.7
|
Gja8
|
gap junction protein, alpha 8 |
| chr9_-_110576192 | 0.47 |
ENSMUST00000199791.2
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr15_+_82230155 | 0.47 |
ENSMUST00000023086.15
|
Smdt1
|
single-pass membrane protein with aspartate rich tail 1 |
| chr18_+_69479211 | 0.46 |
ENSMUST00000201235.4
|
Tcf4
|
transcription factor 4 |
| chr18_-_84104574 | 0.46 |
ENSMUST00000175783.3
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr9_-_110576124 | 0.46 |
ENSMUST00000199862.5
ENSMUST00000198865.5 |
Pth1r
|
parathyroid hormone 1 receptor |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.5 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 2.1 | 6.4 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 2.0 | 6.1 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 1.8 | 5.5 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 1.3 | 5.4 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 1.3 | 4.0 | GO:0036363 | transforming growth factor beta activation(GO:0036363) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 1.2 | 3.7 | GO:0038190 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 1.1 | 3.3 | GO:0046101 | germinal center B cell differentiation(GO:0002314) adenosine catabolic process(GO:0006154) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) purine deoxyribonucleoside metabolic process(GO:0046122) negative regulation of mucus secretion(GO:0070256) |
| 1.1 | 3.2 | GO:0061193 | taste bud development(GO:0061193) |
| 1.0 | 5.1 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 1.0 | 1.0 | GO:2000137 | negative regulation of cell proliferation involved in heart morphogenesis(GO:2000137) |
| 0.9 | 6.2 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.8 | 4.8 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.7 | 8.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.7 | 2.2 | GO:0019049 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.7 | 1.4 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.7 | 2.1 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.7 | 2.0 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.7 | 2.0 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.7 | 3.3 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.6 | 1.8 | GO:1904631 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) response to diterpene(GO:1904629) cellular response to diterpene(GO:1904630) response to glucoside(GO:1904631) cellular response to glucoside(GO:1904632) |
| 0.6 | 1.7 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.5 | 4.9 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.5 | 7.3 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.5 | 2.8 | GO:0051365 | leading edge cell differentiation(GO:0035026) cellular response to potassium ion starvation(GO:0051365) |
| 0.4 | 2.7 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.4 | 5.3 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.4 | 1.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.3 | 1.4 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.3 | 2.7 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.3 | 1.7 | GO:1902608 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.3 | 1.6 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.3 | 2.2 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.3 | 0.9 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.3 | 6.7 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.3 | 3.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.3 | 8.9 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.3 | 4.3 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.2 | 1.9 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.2 | 1.2 | GO:0051885 | positive regulation of anagen(GO:0051885) |
| 0.2 | 1.4 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.2 | 3.8 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.2 | 7.8 | GO:0014823 | response to activity(GO:0014823) |
| 0.2 | 1.8 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.2 | 1.5 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.2 | 1.7 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.2 | 2.9 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.2 | 1.8 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.2 | 1.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 2.4 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.2 | 1.0 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.2 | 4.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.2 | 3.7 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.2 | 3.2 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.1 | 5.1 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.1 | 1.9 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.1 | 1.0 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 5.4 | GO:0060065 | uterus development(GO:0060065) |
| 0.1 | 0.4 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.4 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.1 | 5.8 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 | 0.9 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.1 | 2.0 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 2.5 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.1 | 1.9 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 1.0 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.1 | 2.2 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 2.7 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 1.2 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.1 | 0.4 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.1 | 0.4 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.1 | 0.7 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 1.4 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.1 | 2.3 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 2.1 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 2.4 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.1 | 2.3 | GO:0006356 | regulation of transcription from RNA polymerase I promoter(GO:0006356) |
| 0.1 | 1.1 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.6 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.1 | 5.5 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 0.7 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 1.5 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.1 | 3.7 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 1.7 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.1 | 0.9 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 | 0.3 | GO:0061198 | maintenance of chromatin silencing(GO:0006344) fungiform papilla formation(GO:0061198) |
| 0.0 | 0.4 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 2.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 2.6 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.9 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.5 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 1.0 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 2.7 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.7 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.0 | 3.1 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 1.3 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.3 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.0 | 1.8 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.4 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.0 | 7.3 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 3.4 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 2.5 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 3.1 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 0.0 | 2.1 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 3.7 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 13.8 | GO:0030855 | epithelial cell differentiation(GO:0030855) |
| 0.0 | 0.7 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.7 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 3.8 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 1.3 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.5 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 2.5 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 1.2 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 0.2 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 3.7 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 8.9 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 1.0 | 5.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.9 | 3.7 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.8 | 9.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.7 | 4.9 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.6 | 1.9 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.6 | 2.8 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.4 | 2.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.3 | 4.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.2 | 6.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 5.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.2 | 1.6 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 0.7 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 3.9 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 1.8 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 15.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 3.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 5.7 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.7 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 4.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 3.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 3.8 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 12.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 1.7 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 0.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.7 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.6 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 1.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 1.0 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 2.1 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 0.4 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 1.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 3.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 4.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.6 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 2.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 6.5 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 2.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 6.9 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 1.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 18.6 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 3.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 5.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.3 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 8.9 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.9 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 21.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 5.9 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 9.4 | GO:0050436 | microfibril binding(GO:0050436) |
| 1.5 | 10.3 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 1.5 | 8.8 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 1.4 | 4.2 | GO:0035375 | zymogen binding(GO:0035375) |
| 1.2 | 3.7 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 1.2 | 3.7 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 1.1 | 5.5 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) bradykinin receptor binding(GO:0031711) |
| 0.7 | 6.7 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.7 | 6.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.7 | 2.0 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 0.6 | 5.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.5 | 2.5 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.4 | 4.8 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.4 | 3.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.4 | 6.1 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.4 | 2.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.3 | 8.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.3 | 1.7 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.3 | 5.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 6.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 3.3 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.2 | 1.5 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.2 | 1.4 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.2 | 2.6 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.2 | 5.5 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.2 | 1.0 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.2 | 3.7 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.2 | 5.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 1.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 4.0 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 3.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.7 | GO:0030977 | taurine binding(GO:0030977) |
| 0.1 | 2.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 1.2 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 0.9 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 1.7 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 3.2 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 2.0 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.8 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 2.4 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 0.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 2.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 2.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 0.3 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.1 | 1.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 26.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 0.3 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.1 | 1.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 1.5 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.8 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.1 | 2.0 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 2.1 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.9 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 1.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.3 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.7 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 5.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.5 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 6.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 3.9 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 6.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 1.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 1.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.2 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 1.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.4 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 3.3 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.2 | GO:0034061 | DNA polymerase activity(GO:0034061) |
| 0.0 | 0.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 5.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 3.2 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 1.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 1.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.6 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 3.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 6.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 3.7 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.1 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.4 | 3.7 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 2.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 5.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 6.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 7.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.8 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 4.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 8.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 6.0 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 5.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 5.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.7 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 20.8 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 19.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 17.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 1.7 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 2.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 16.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 3.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.7 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 2.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.8 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 2.4 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 8.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.4 | 5.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.4 | 3.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.3 | 10.3 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.2 | 8.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 2.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 3.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 6.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.0 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 3.9 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 1.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 2.0 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 4.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 6.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 3.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 7.5 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 3.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.8 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 1.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 1.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 2.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 2.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.7 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 1.9 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |