Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Sox1
Z-value: 0.55
Transcription factors associated with Sox1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox1
|
ENSMUSG00000096014.2 | Sox1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox1 | mm39_v1_chr8_+_12445287_12445295 | -0.78 | 8.9e-16 | Click! |
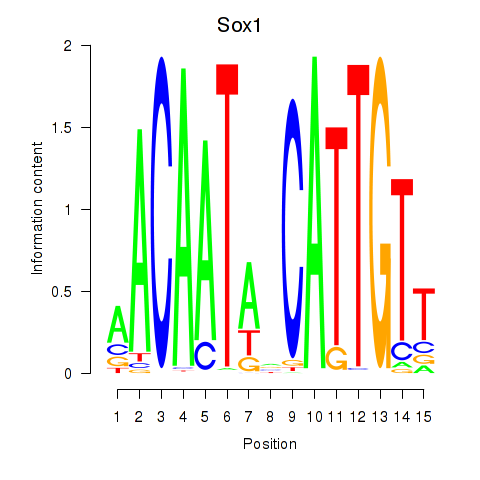
Activity profile of Sox1 motif
Sorted Z-values of Sox1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_34482183 | 7.71 |
ENSMUST00000040828.7
ENSMUST00000237342.2 ENSMUST00000237866.2 |
H2-Ab1
|
histocompatibility 2, class II antigen A, beta 1 |
| chr17_-_34506744 | 7.23 |
ENSMUST00000174751.2
ENSMUST00000040655.14 |
H2-Aa
|
histocompatibility 2, class II antigen A, alpha |
| chr14_+_65903840 | 3.42 |
ENSMUST00000022610.15
|
Scara5
|
scavenger receptor class A, member 5 |
| chr1_-_173703424 | 3.36 |
ENSMUST00000186442.7
|
Mndal
|
myeloid nuclear differentiation antigen like |
| chr9_-_114219685 | 3.19 |
ENSMUST00000084881.5
|
Crtap
|
cartilage associated protein |
| chr17_+_48047955 | 2.73 |
ENSMUST00000086932.10
|
Tfeb
|
transcription factor EB |
| chr2_+_84629172 | 2.70 |
ENSMUST00000102642.9
ENSMUST00000150325.2 |
Ube2l6
|
ubiquitin-conjugating enzyme E2L 6 |
| chr6_+_37507108 | 2.56 |
ENSMUST00000040987.11
|
Akr1d1
|
aldo-keto reductase family 1, member D1 |
| chr15_-_74869483 | 2.51 |
ENSMUST00000023248.13
|
Ly6a
|
lymphocyte antigen 6 complex, locus A |
| chr15_-_74869684 | 2.14 |
ENSMUST00000190188.2
ENSMUST00000189068.7 ENSMUST00000186526.7 ENSMUST00000187171.2 ENSMUST00000187994.7 |
Ly6a
|
lymphocyte antigen 6 complex, locus A |
| chr14_+_65903878 | 2.13 |
ENSMUST00000069226.7
|
Scara5
|
scavenger receptor class A, member 5 |
| chr5_+_21391282 | 2.11 |
ENSMUST00000036031.13
ENSMUST00000198937.2 |
Gsap
|
gamma-secretase activating protein |
| chr6_-_52195663 | 2.09 |
ENSMUST00000134367.4
|
Hoxa7
|
homeobox A7 |
| chr1_+_171723231 | 1.90 |
ENSMUST00000097466.3
|
Gm10521
|
predicted gene 10521 |
| chr4_-_131664478 | 1.85 |
ENSMUST00000155990.8
|
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr7_+_67305162 | 1.79 |
ENSMUST00000107470.2
|
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr2_-_84481101 | 1.74 |
ENSMUST00000111691.2
|
Ctnnd1
|
catenin (cadherin associated protein), delta 1 |
| chr14_-_56339915 | 1.58 |
ENSMUST00000015583.2
|
Ctsg
|
cathepsin G |
| chr2_-_84481058 | 1.54 |
ENSMUST00000111670.9
ENSMUST00000111697.9 ENSMUST00000111696.8 ENSMUST00000111678.8 ENSMUST00000111690.8 ENSMUST00000111695.8 ENSMUST00000111677.8 ENSMUST00000111698.8 ENSMUST00000099941.9 ENSMUST00000111676.8 ENSMUST00000111694.8 ENSMUST00000111675.8 ENSMUST00000111689.8 ENSMUST00000111687.8 ENSMUST00000111692.8 ENSMUST00000111685.8 ENSMUST00000111686.8 ENSMUST00000111688.8 ENSMUST00000111693.8 ENSMUST00000111684.8 |
Ctnnd1
|
catenin (cadherin associated protein), delta 1 |
| chr19_+_18818001 | 1.53 |
ENSMUST00000237020.2
|
Trpm6
|
transient receptor potential cation channel, subfamily M, member 6 |
| chr11_-_69768875 | 1.49 |
ENSMUST00000178597.3
|
Tmem95
|
transmembrane protein 95 |
| chr1_-_194813843 | 1.42 |
ENSMUST00000075451.12
ENSMUST00000191775.2 |
Cr1l
|
complement component (3b/4b) receptor 1-like |
| chr19_-_8382424 | 1.40 |
ENSMUST00000064507.12
ENSMUST00000120540.2 ENSMUST00000096269.11 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr11_+_96024612 | 1.36 |
ENSMUST00000167258.8
|
Ttll6
|
tubulin tyrosine ligase-like family, member 6 |
| chrX_+_100473161 | 1.30 |
ENSMUST00000033673.7
|
Nono
|
non-POU-domain-containing, octamer binding protein |
| chr13_+_42454922 | 1.14 |
ENSMUST00000021796.9
|
Edn1
|
endothelin 1 |
| chr12_+_52144511 | 1.13 |
ENSMUST00000040090.16
|
Nubpl
|
nucleotide binding protein-like |
| chr7_+_119289249 | 1.05 |
ENSMUST00000047045.10
|
Acsm4
|
acyl-CoA synthetase medium-chain family member 4 |
| chr1_-_194813631 | 0.94 |
ENSMUST00000194111.6
ENSMUST00000193094.6 |
Cr1l
|
complement component (3b/4b) receptor 1-like |
| chr9_+_44238089 | 0.88 |
ENSMUST00000054708.5
|
Dpagt1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr3_+_90497554 | 0.82 |
ENSMUST00000179550.2
|
S100a2
|
S100 calcium binding protein A2 |
| chr3_+_60503051 | 0.81 |
ENSMUST00000192757.6
ENSMUST00000193518.6 ENSMUST00000195817.3 |
Mbnl1
|
muscleblind like splicing factor 1 |
| chr6_+_70170212 | 0.74 |
ENSMUST00000103382.2
|
Igkv8-26
|
immunoglobulin kappa variable 8-26 |
| chr6_+_70332836 | 0.74 |
ENSMUST00000103390.3
|
Igkv8-18
|
immunoglobulin kappa variable 8-18 |
| chr9_+_123596276 | 0.72 |
ENSMUST00000166236.9
ENSMUST00000111454.4 ENSMUST00000168910.2 |
Ccr9
|
chemokine (C-C motif) receptor 9 |
| chr8_-_89362745 | 0.71 |
ENSMUST00000034087.9
|
Snx20
|
sorting nexin 20 |
| chr2_+_87185159 | 0.65 |
ENSMUST00000215163.3
|
Olfr1120
|
olfactory receptor 1120 |
| chr15_-_74920518 | 0.62 |
ENSMUST00000185372.2
ENSMUST00000187347.7 ENSMUST00000188845.7 ENSMUST00000185200.7 ENSMUST00000179762.8 ENSMUST00000191216.7 ENSMUST00000065408.16 |
Ly6c1
|
lymphocyte antigen 6 complex, locus C1 |
| chr11_+_73244561 | 0.57 |
ENSMUST00000108465.4
|
Olfr20
|
olfactory receptor 20 |
| chrX_+_162945162 | 0.55 |
ENSMUST00000131543.2
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr7_+_13357892 | 0.54 |
ENSMUST00000108525.4
|
Sult2a5
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 5 |
| chr4_-_120604445 | 0.53 |
ENSMUST00000030376.8
|
Kcnq4
|
potassium voltage-gated channel, subfamily Q, member 4 |
| chr5_+_21942139 | 0.51 |
ENSMUST00000030882.12
|
Pmpcb
|
peptidase (mitochondrial processing) beta |
| chr11_+_96025045 | 0.48 |
ENSMUST00000107680.2
|
Ttll6
|
tubulin tyrosine ligase-like family, member 6 |
| chr14_+_54032814 | 0.47 |
ENSMUST00000103671.4
|
Trav13-5
|
T cell receptor alpha variable 13-5 |
| chr7_+_103620359 | 0.45 |
ENSMUST00000209473.4
|
Olfr635
|
olfactory receptor 635 |
| chr7_+_79992839 | 0.44 |
ENSMUST00000032747.7
ENSMUST00000206480.2 ENSMUST00000206074.2 ENSMUST00000206122.2 |
Hddc3
|
HD domain containing 3 |
| chr9_+_123921573 | 0.43 |
ENSMUST00000111442.3
ENSMUST00000171499.3 |
Ccr5
|
chemokine (C-C motif) receptor 5 |
| chr2_-_34645241 | 0.42 |
ENSMUST00000102800.9
|
Gapvd1
|
GTPase activating protein and VPS9 domains 1 |
| chr3_+_32490300 | 0.40 |
ENSMUST00000029201.14
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr9_-_108183140 | 0.40 |
ENSMUST00000195615.2
|
Tcta
|
T cell leukemia translocation altered gene |
| chr16_-_37474772 | 0.39 |
ENSMUST00000023514.4
|
Ndufb4
|
NADH:ubiquinone oxidoreductase subunit B4 |
| chr4_+_63478478 | 0.37 |
ENSMUST00000080336.4
|
Tmem268
|
transmembrane protein 268 |
| chr2_+_87726521 | 0.37 |
ENSMUST00000052300.2
|
Olfr1153
|
olfactory receptor 1153 |
| chr5_-_82272549 | 0.36 |
ENSMUST00000188072.2
ENSMUST00000185410.2 |
1700031L13Rik
|
RIKEN cDNA 1700031L13 gene |
| chr17_+_35188888 | 0.34 |
ENSMUST00000173680.2
|
Gm20481
|
predicted gene 20481 |
| chr17_-_21134625 | 0.21 |
ENSMUST00000077001.3
|
Vmn1r232
|
vomeronasal 1 receptor 232 |
| chr14_-_35533922 | 0.20 |
ENSMUST00000043266.7
|
4930596D02Rik
|
RIKEN cDNA 4930596D02 gene |
| chr2_+_34296783 | 0.19 |
ENSMUST00000149383.8
ENSMUST00000124443.8 ENSMUST00000141253.2 ENSMUST00000113124.8 |
Mapkap1
|
mitogen-activated protein kinase associated protein 1 |
| chr7_-_119744509 | 0.17 |
ENSMUST00000208874.2
ENSMUST00000033207.6 |
Zp2
|
zona pellucida glycoprotein 2 |
| chr6_-_66757618 | 0.17 |
ENSMUST00000227493.2
|
Vmn1r38
|
vomeronasal 1 receptor 38 |
| chr8_+_13209141 | 0.16 |
ENSMUST00000033824.8
|
Lamp1
|
lysosomal-associated membrane protein 1 |
| chr6_+_121160626 | 0.13 |
ENSMUST00000118234.8
ENSMUST00000088561.10 ENSMUST00000137432.8 ENSMUST00000120066.8 |
Pex26
|
peroxisomal biogenesis factor 26 |
| chr2_+_88644840 | 0.11 |
ENSMUST00000214703.2
|
Olfr1202
|
olfactory receptor 1202 |
| chr11_+_49355873 | 0.11 |
ENSMUST00000213674.2
ENSMUST00000204518.3 |
Olfr1386
|
olfactory receptor 1386 |
| chr18_-_70409277 | 0.08 |
ENSMUST00000239144.2
|
Gm36255
|
predicted gene, 36255 |
| chr7_-_103710652 | 0.05 |
ENSMUST00000074064.5
|
4930516K23Rik
|
RIKEN cDNA 4930516K23 gene |
| chr6_+_114625771 | 0.04 |
ENSMUST00000182510.8
|
Atg7
|
autophagy related 7 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.7 | GO:0002343 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 1.1 | 3.2 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.9 | 2.6 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.7 | 2.7 | GO:0009816 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.5 | 3.3 | GO:0060690 | epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.4 | 1.1 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.4 | 7.2 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.3 | 2.7 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.3 | 1.8 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.2 | 0.7 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.2 | 2.4 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.2 | 5.5 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.2 | 2.1 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.2 | 1.6 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 0.1 | 1.3 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 2.1 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.1 | 0.5 | GO:0015827 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.1 | 0.4 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 0.9 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 1.8 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 1.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.4 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 0.5 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.1 | 0.2 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.5 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 1.5 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.4 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.2 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.0 | 0.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 3.4 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 14.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 3.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.2 | 1.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 0.4 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 1.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.2 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.0 | 0.5 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 2.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.8 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 5.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 1.8 | GO:0036064 | ciliary basal body(GO:0036064) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.5 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 0.5 | 2.6 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.5 | 7.4 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.5 | 2.7 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.3 | 1.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.2 | 0.9 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.2 | 7.2 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.2 | 1.8 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.1 | 1.0 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.1 | 0.5 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 1.3 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.8 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 1.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 1.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.4 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 2.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.1 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 4.7 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 0.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 1.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.1 | PID ENDOTHELIN PATHWAY | Endothelins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 0.4 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 1.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 2.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 3.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.9 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.6 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |