Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Sox11
Z-value: 0.42
Transcription factors associated with Sox11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox11
|
ENSMUSG00000063632.7 | Sox11 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox11 | mm39_v1_chr12_-_27392356_27392573 | 0.38 | 1.0e-03 | Click! |
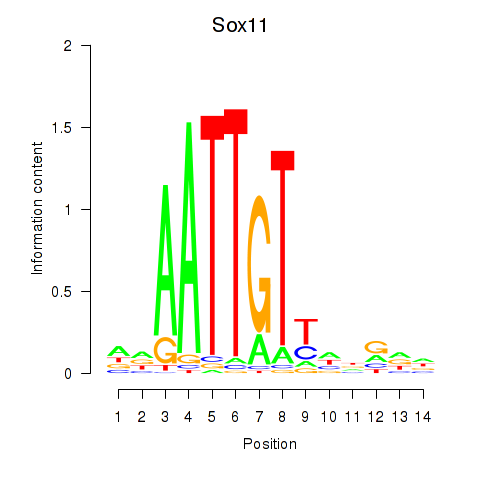
Activity profile of Sox11 motif
Sorted Z-values of Sox11 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox11
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_60806810 | 3.25 |
ENSMUST00000163779.8
|
Snca
|
synuclein, alpha |
| chr11_-_116572116 | 2.25 |
ENSMUST00000144398.2
|
St6galnac2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2 |
| chr13_+_4109566 | 1.78 |
ENSMUST00000041768.7
|
Akr1c14
|
aldo-keto reductase family 1, member C14 |
| chrX_+_139808351 | 1.75 |
ENSMUST00000033806.5
|
Vsig1
|
V-set and immunoglobulin domain containing 1 |
| chr6_-_42670021 | 1.47 |
ENSMUST00000121083.8
|
Tcaf1
|
TRPM8 channel-associated factor 1 |
| chr5_+_14564932 | 1.43 |
ENSMUST00000182407.8
ENSMUST00000030691.17 |
Pclo
|
piccolo (presynaptic cytomatrix protein) |
| chr5_+_89175815 | 1.39 |
ENSMUST00000130041.8
|
Slc4a4
|
solute carrier family 4 (anion exchanger), member 4 |
| chr5_+_89175894 | 1.34 |
ENSMUST00000113216.9
ENSMUST00000134303.2 |
Slc4a4
|
solute carrier family 4 (anion exchanger), member 4 |
| chr6_-_42669963 | 1.31 |
ENSMUST00000045140.5
|
Tcaf1
|
TRPM8 channel-associated factor 1 |
| chr13_-_99653045 | 1.28 |
ENSMUST00000064762.6
|
Map1b
|
microtubule-associated protein 1B |
| chr2_+_154390808 | 1.27 |
ENSMUST00000045116.11
ENSMUST00000109709.4 |
1700003F12Rik
|
RIKEN cDNA 1700003F12 gene |
| chrX_+_162923474 | 1.21 |
ENSMUST00000073973.11
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr11_-_41891662 | 1.11 |
ENSMUST00000070725.11
|
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
| chr10_+_5589210 | 1.11 |
ENSMUST00000019906.6
|
Vip
|
vasoactive intestinal polypeptide |
| chr10_+_34359395 | 1.08 |
ENSMUST00000019913.15
|
Frk
|
fyn-related kinase |
| chr10_+_34359513 | 1.03 |
ENSMUST00000170771.3
|
Frk
|
fyn-related kinase |
| chr1_-_149798090 | 1.02 |
ENSMUST00000111926.9
|
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr7_+_81824544 | 0.97 |
ENSMUST00000032874.14
|
Sh3gl3
|
SH3-domain GRB2-like 3 |
| chr2_-_90301592 | 0.96 |
ENSMUST00000111493.8
|
Ptprj
|
protein tyrosine phosphatase, receptor type, J |
| chr6_-_136150491 | 0.96 |
ENSMUST00000111905.8
ENSMUST00000152012.8 ENSMUST00000143943.8 ENSMUST00000125905.2 |
Grin2b
|
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
| chr18_+_37143758 | 0.89 |
ENSMUST00000115657.10
ENSMUST00000192447.6 |
Pcdha11
|
protocadherin alpha 11 |
| chr5_+_8096074 | 0.86 |
ENSMUST00000088786.11
|
Sri
|
sorcin |
| chr17_+_43327412 | 0.86 |
ENSMUST00000024708.6
|
Tnfrsf21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr3_-_102074246 | 0.84 |
ENSMUST00000161021.2
|
Vangl1
|
VANGL planar cell polarity 1 |
| chr2_+_3115250 | 0.81 |
ENSMUST00000072955.12
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr1_+_177557380 | 0.81 |
ENSMUST00000016106.6
|
1700016C15Rik
|
RIKEN cDNA 1700016C15 gene |
| chrX_+_162945162 | 0.77 |
ENSMUST00000131543.2
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr2_-_110591909 | 0.72 |
ENSMUST00000140777.2
|
Ano3
|
anoctamin 3 |
| chr9_-_16289527 | 0.66 |
ENSMUST00000082170.6
|
Fat3
|
FAT atypical cadherin 3 |
| chr9_+_48073296 | 0.66 |
ENSMUST00000216998.2
ENSMUST00000215780.2 |
Nxpe4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr4_+_135455427 | 0.65 |
ENSMUST00000102546.4
|
Il22ra1
|
interleukin 22 receptor, alpha 1 |
| chr19_-_4675352 | 0.62 |
ENSMUST00000224707.2
ENSMUST00000237059.2 |
Rce1
|
Ras converting CAAX endopeptidase 1 |
| chr9_-_51013361 | 0.61 |
ENSMUST00000170947.3
|
Hoatz
|
HOATZ cilia and flagella associated protein |
| chr6_-_40496403 | 0.60 |
ENSMUST00000031967.6
|
Prss37
|
protease, serine 37 |
| chr9_-_29323032 | 0.59 |
ENSMUST00000115236.2
|
Ntm
|
neurotrimin |
| chr14_-_121935829 | 0.59 |
ENSMUST00000040700.9
ENSMUST00000212181.2 |
Dock9
|
dedicator of cytokinesis 9 |
| chr18_+_37106851 | 0.58 |
ENSMUST00000192631.2
|
Pcdha7
|
protocadherin alpha 7 |
| chr3_+_133942244 | 0.55 |
ENSMUST00000181904.3
|
Cxxc4
|
CXXC finger 4 |
| chr18_-_35841435 | 0.55 |
ENSMUST00000236738.2
ENSMUST00000237995.2 |
Dnajc18
|
DnaJ heat shock protein family (Hsp40) member C18 |
| chr6_-_87312743 | 0.54 |
ENSMUST00000042025.12
ENSMUST00000205033.2 |
Antxr1
|
anthrax toxin receptor 1 |
| chr17_+_35827997 | 0.51 |
ENSMUST00000164242.9
ENSMUST00000045956.14 |
Cchcr1
|
coiled-coil alpha-helical rod protein 1 |
| chr9_-_58648826 | 0.49 |
ENSMUST00000098674.6
|
Rec114
|
REC114 meiotic recombination protein |
| chr19_-_4675300 | 0.49 |
ENSMUST00000225264.2
ENSMUST00000237022.2 ENSMUST00000224675.3 |
Rce1
|
Ras converting CAAX endopeptidase 1 |
| chr10_-_8632519 | 0.48 |
ENSMUST00000212869.2
|
Sash1
|
SAM and SH3 domain containing 1 |
| chr19_+_8815024 | 0.48 |
ENSMUST00000159634.8
|
Bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr19_-_46958001 | 0.47 |
ENSMUST00000235234.2
|
Nt5c2
|
5'-nucleotidase, cytosolic II |
| chr10_-_8638215 | 0.46 |
ENSMUST00000212553.2
|
Sash1
|
SAM and SH3 domain containing 1 |
| chr13_+_24925192 | 0.46 |
ENSMUST00000136906.2
|
Armh2
|
armadillo-like helical domain containing 2 |
| chr11_+_4823951 | 0.45 |
ENSMUST00000038570.9
|
Nipsnap1
|
nipsnap homolog 1 |
| chr11_-_33942981 | 0.45 |
ENSMUST00000238903.2
|
Kcnip1
|
Kv channel-interacting protein 1 |
| chr2_-_79287095 | 0.44 |
ENSMUST00000041099.5
|
Neurod1
|
neurogenic differentiation 1 |
| chr10_+_90412114 | 0.44 |
ENSMUST00000182427.8
ENSMUST00000182053.8 ENSMUST00000182113.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr5_+_145020640 | 0.44 |
ENSMUST00000031625.15
|
Arpc1a
|
actin related protein 2/3 complex, subunit 1A |
| chr6_-_87312681 | 0.42 |
ENSMUST00000204805.3
|
Antxr1
|
anthrax toxin receptor 1 |
| chr10_+_119828821 | 0.40 |
ENSMUST00000105261.9
|
Grip1
|
glutamate receptor interacting protein 1 |
| chr4_+_152160713 | 0.40 |
ENSMUST00000239025.2
|
Plekhg5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chrX_-_100103249 | 0.39 |
ENSMUST00000113718.8
ENSMUST00000113716.3 |
Tex11
|
testis expressed gene 11 |
| chr9_+_45029080 | 0.39 |
ENSMUST00000170998.9
ENSMUST00000093855.4 |
Scn2b
|
sodium channel, voltage-gated, type II, beta |
| chr5_+_144037171 | 0.39 |
ENSMUST00000041804.8
|
Lmtk2
|
lemur tyrosine kinase 2 |
| chr4_-_46404224 | 0.38 |
ENSMUST00000107764.9
|
Hemgn
|
hemogen |
| chr13_-_41640757 | 0.37 |
ENSMUST00000021794.14
|
Nedd9
|
neural precursor cell expressed, developmentally down-regulated gene 9 |
| chr8_+_106154266 | 0.37 |
ENSMUST00000067305.8
|
Lrrc36
|
leucine rich repeat containing 36 |
| chr18_+_37580692 | 0.36 |
ENSMUST00000052387.5
|
Pcdhb14
|
protocadherin beta 14 |
| chr16_-_50151350 | 0.35 |
ENSMUST00000114488.8
|
Bbx
|
bobby sox HMG box containing |
| chr6_-_69394425 | 0.32 |
ENSMUST00000199160.2
|
Igkv4-61
|
immunoglobulin kappa chain variable 4-61 |
| chrX_-_100103220 | 0.31 |
ENSMUST00000009814.10
|
Tex11
|
testis expressed gene 11 |
| chr5_-_148489457 | 0.31 |
ENSMUST00000079324.14
|
Ubl3
|
ubiquitin-like 3 |
| chr18_-_75830595 | 0.30 |
ENSMUST00000165559.3
|
Ctif
|
CBP80/20-dependent translation initiation factor |
| chr3_+_28859585 | 0.30 |
ENSMUST00000043867.11
ENSMUST00000194649.2 |
Rpl22l1
|
ribosomal protein L22 like 1 |
| chrX_+_84617624 | 0.27 |
ENSMUST00000048250.10
ENSMUST00000137438.2 ENSMUST00000146063.2 |
Tab3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr10_+_97482946 | 0.27 |
ENSMUST00000105285.4
|
Epyc
|
epiphycan |
| chr5_+_16758897 | 0.27 |
ENSMUST00000196645.2
|
Hgf
|
hepatocyte growth factor |
| chr4_-_129271909 | 0.26 |
ENSMUST00000030610.3
|
Zbtb8a
|
zinc finger and BTB domain containing 8a |
| chr2_+_121991181 | 0.26 |
ENSMUST00000036089.8
|
Trim69
|
tripartite motif-containing 69 |
| chr5_+_145020910 | 0.25 |
ENSMUST00000124379.3
|
Arpc1a
|
actin related protein 2/3 complex, subunit 1A |
| chr10_+_119828867 | 0.24 |
ENSMUST00000154238.8
|
Grip1
|
glutamate receptor interacting protein 1 |
| chr2_+_111329683 | 0.23 |
ENSMUST00000219064.3
|
Olfr1291-ps1
|
olfactory receptor 1291, pseudogene 1 |
| chr2_-_86109346 | 0.21 |
ENSMUST00000217294.2
ENSMUST00000217245.2 ENSMUST00000216432.2 |
Olfr1051
|
olfactory receptor 1051 |
| chr8_+_106052970 | 0.21 |
ENSMUST00000015000.12
ENSMUST00000098453.9 |
Tmem208
|
transmembrane protein 208 |
| chr5_-_148489593 | 0.21 |
ENSMUST00000201595.4
ENSMUST00000164904.2 |
Ubl3
|
ubiquitin-like 3 |
| chr16_-_17729167 | 0.20 |
ENSMUST00000232423.2
ENSMUST00000003621.10 |
Ess2
|
ess-2 splicing factor |
| chr4_+_109092459 | 0.20 |
ENSMUST00000106631.9
|
Calr4
|
calreticulin 4 |
| chr11_-_106084334 | 0.20 |
ENSMUST00000007444.14
ENSMUST00000152008.2 ENSMUST00000103072.10 ENSMUST00000106867.2 |
Strada
|
STE20-related kinase adaptor alpha |
| chrX_-_50889955 | 0.20 |
ENSMUST00000069509.4
ENSMUST00000114869.8 |
Usp26
|
ubiquitin specific peptidase 26 |
| chr8_-_35432783 | 0.17 |
ENSMUST00000033929.6
|
Tnks
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr1_-_171122509 | 0.17 |
ENSMUST00000111302.4
ENSMUST00000080001.9 |
Ufc1
|
ubiquitin-fold modifier conjugating enzyme 1 |
| chr7_+_105053775 | 0.16 |
ENSMUST00000033187.6
ENSMUST00000210344.2 |
Cnga4
|
cyclic nucleotide gated channel alpha 4 |
| chr4_-_149858694 | 0.15 |
ENSMUST00000105686.3
|
Slc25a33
|
solute carrier family 25, member 33 |
| chr6_-_48817389 | 0.15 |
ENSMUST00000204783.3
|
Tmem176b
|
transmembrane protein 176B |
| chr7_-_85985625 | 0.15 |
ENSMUST00000069279.5
|
Olfr307
|
olfactory receptor 307 |
| chr9_-_19968858 | 0.14 |
ENSMUST00000216538.2
ENSMUST00000212098.3 |
Olfr867
|
olfactory receptor 867 |
| chr6_-_69584812 | 0.14 |
ENSMUST00000103359.3
|
Igkv4-55
|
immunoglobulin kappa variable 4-55 |
| chr10_+_121200984 | 0.13 |
ENSMUST00000040344.7
|
Gns
|
glucosamine (N-acetyl)-6-sulfatase |
| chr6_-_48817675 | 0.13 |
ENSMUST00000203265.3
ENSMUST00000205159.3 |
Tmem176b
|
transmembrane protein 176B |
| chr13_-_22689551 | 0.13 |
ENSMUST00000228020.2
|
Vmn1r202
|
vomeronasal 1 receptor 202 |
| chr2_-_111100733 | 0.12 |
ENSMUST00000099619.6
|
Olfr1277
|
olfactory receptor 1277 |
| chr6_-_119940694 | 0.12 |
ENSMUST00000161512.3
|
Wnk1
|
WNK lysine deficient protein kinase 1 |
| chr4_+_48124902 | 0.12 |
ENSMUST00000107721.8
ENSMUST00000153502.8 ENSMUST00000107720.9 ENSMUST00000064765.6 |
Stx17
|
syntaxin 17 |
| chr18_+_44403169 | 0.12 |
ENSMUST00000042747.4
|
Npy6r
|
neuropeptide Y receptor Y6 |
| chr6_-_48817529 | 0.11 |
ENSMUST00000203501.2
|
Tmem176b
|
transmembrane protein 176B |
| chr6_-_8180174 | 0.10 |
ENSMUST00000213284.2
|
Col28a1
|
collagen, type XXVIII, alpha 1 |
| chr6_+_108771840 | 0.10 |
ENSMUST00000204483.2
|
Arl8b
|
ADP-ribosylation factor-like 8B |
| chr3_-_10262275 | 0.10 |
ENSMUST00000193487.6
ENSMUST00000029038.8 ENSMUST00000191678.6 |
Fabp9
|
fatty acid binding protein 9, testis |
| chr8_-_62576140 | 0.10 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr6_-_69020489 | 0.09 |
ENSMUST00000103342.4
|
Igkv4-79
|
immunoglobulin kappa variable 4-79 |
| chr5_-_86893645 | 0.09 |
ENSMUST00000161306.2
|
Tmprss11e
|
transmembrane protease, serine 11e |
| chr7_-_55612213 | 0.09 |
ENSMUST00000032635.14
ENSMUST00000152649.8 |
Nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 homolog (human) |
| chr16_-_38162174 | 0.08 |
ENSMUST00000114740.3
|
Cfap91
|
cilia and flagella associated protein 91 |
| chr4_+_59003121 | 0.07 |
ENSMUST00000095070.4
ENSMUST00000174664.2 |
Dnajc25
Gm20503
|
DnaJ heat shock protein family (Hsp40) member C25 predicted gene 20503 |
| chr1_+_44159106 | 0.07 |
ENSMUST00000114709.3
ENSMUST00000129068.2 |
Bivm
|
basic, immunoglobulin-like variable motif containing |
| chr15_-_5137951 | 0.06 |
ENSMUST00000141020.2
|
Card6
|
caspase recruitment domain family, member 6 |
| chr13_-_113755082 | 0.06 |
ENSMUST00000109241.5
|
Snx18
|
sorting nexin 18 |
| chr12_+_8971603 | 0.05 |
ENSMUST00000020909.4
|
Laptm4a
|
lysosomal-associated protein transmembrane 4A |
| chr10_+_115405891 | 0.04 |
ENSMUST00000173620.2
|
A930009A15Rik
|
RIKEN cDNA A930009A15 gene |
| chr14_-_20438890 | 0.04 |
ENSMUST00000022345.7
|
Dnajc9
|
DnaJ heat shock protein family (Hsp40) member C9 |
| chrM_+_7779 | 0.04 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr4_+_109092610 | 0.03 |
ENSMUST00000106628.8
|
Calr4
|
calreticulin 4 |
| chr2_-_86589298 | 0.02 |
ENSMUST00000215991.2
ENSMUST00000217043.3 |
Olfr1090
|
olfactory receptor 1090 |
| chr13_-_95615229 | 0.02 |
ENSMUST00000022186.5
|
S100z
|
S100 calcium binding protein, zeta |
| chr10_-_75946790 | 0.02 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr4_-_97472844 | 0.02 |
ENSMUST00000107067.8
ENSMUST00000107068.9 |
E130114P18Rik
|
RIKEN cDNA E130114P18 gene |
| chr6_-_69220672 | 0.02 |
ENSMUST00000196201.2
|
Igkv4-71
|
immunoglobulin kappa chain variable 4-71 |
| chr5_-_86666408 | 0.02 |
ENSMUST00000140095.2
ENSMUST00000134179.8 |
Tmprss11g
|
transmembrane protease, serine 11g |
| chr8_+_85952617 | 0.02 |
ENSMUST00000218663.2
|
Olfr371
|
olfactory receptor 371 |
| chr17_-_21125353 | 0.01 |
ENSMUST00000235948.2
ENSMUST00000238096.2 |
Vmn1r231
|
vomeronasal 1 receptor 231 |
| chr14_+_54406112 | 0.00 |
ENSMUST00000103694.3
ENSMUST00000199452.2 |
Traj49
|
T cell receptor alpha joining 49 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.5 | 2.0 | GO:0015801 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.5 | 1.4 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.3 | 2.8 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.3 | 2.2 | GO:0097503 | sialylation(GO:0097503) |
| 0.3 | 1.1 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.2 | 0.9 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.2 | 1.0 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.2 | 0.9 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.2 | 1.0 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 | 1.0 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.7 | GO:0035822 | meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
| 0.1 | 2.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 2.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 1.1 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.1 | 0.9 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.1 | 0.7 | GO:0061590 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.1 | 1.0 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.4 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.1 | 1.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.1 | 1.3 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) |
| 0.1 | 0.2 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.1 | 0.2 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 1.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.4 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.2 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.2 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.4 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.1 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 0.4 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.3 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 2.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.7 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.4 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.0 | 1.2 | GO:0009988 | cell-cell recognition(GO:0009988) |
| 0.0 | 0.7 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.4 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.5 | 1.4 | GO:0044317 | rod spherule(GO:0044317) |
| 0.2 | 0.9 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 0.7 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.7 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 1.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 1.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 1.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.0 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.2 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 1.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 2.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 4.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.3 | 1.8 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.3 | 3.2 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.2 | 0.7 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 1.3 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 2.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 1.0 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 1.0 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 1.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.4 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 2.2 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0005223 | intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.9 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 2.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.6 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 1.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 3.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.5 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.0 | 0.9 | GO:0032947 | protein complex scaffold(GO:0032947) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 2.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.0 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.2 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.0 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 1.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.3 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 1.1 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 2.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |