Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
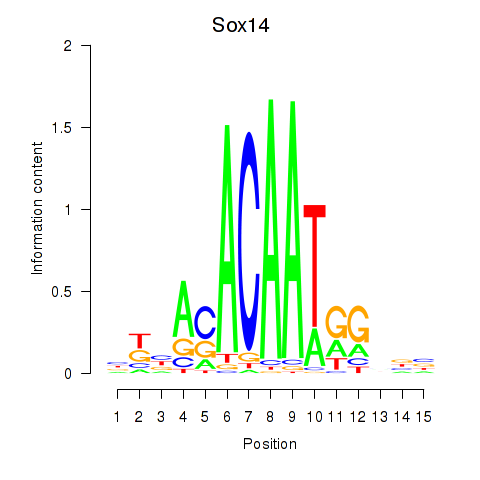
Results for Sox14
Z-value: 1.16
Transcription factors associated with Sox14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox14
|
ENSMUSG00000053747.10 | Sox14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox14 | mm39_v1_chr9_-_99758200_99758245 | -0.26 | 2.9e-02 | Click! |
Activity profile of Sox14 motif
Sorted Z-values of Sox14 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox14
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_68893253 | 10.80 |
ENSMUST00000225767.3
ENSMUST00000111072.8 ENSMUST00000022642.6 ENSMUST00000224039.2 |
Adam28
|
a disintegrin and metallopeptidase domain 28 |
| chr1_-_79838897 | 7.05 |
ENSMUST00000190724.2
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chr17_+_34457868 | 6.59 |
ENSMUST00000095342.11
ENSMUST00000167280.8 ENSMUST00000236838.2 |
H2-Ob
|
histocompatibility 2, O region beta locus |
| chr6_+_70700207 | 6.39 |
ENSMUST00000103407.3
ENSMUST00000199487.2 |
Igkj3
|
immunoglobulin kappa joining 3 |
| chr9_+_22010512 | 6.24 |
ENSMUST00000214601.2
ENSMUST00000001384.6 |
Cnn1
|
calponin 1 |
| chr12_-_115122455 | 5.78 |
ENSMUST00000103523.2
|
Ighv1-53
|
immunoglobulin heavy variable 1-53 |
| chr1_+_134110142 | 5.76 |
ENSMUST00000082060.10
ENSMUST00000153856.8 ENSMUST00000133701.8 ENSMUST00000132873.8 |
Chil1
|
chitinase-like 1 |
| chr11_+_68936457 | 5.40 |
ENSMUST00000108666.8
ENSMUST00000021277.6 |
Aurkb
|
aurora kinase B |
| chrX_-_165992145 | 5.28 |
ENSMUST00000112176.8
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr14_-_40907106 | 5.21 |
ENSMUST00000077136.5
|
Sftpd
|
surfactant associated protein D |
| chr1_+_134109888 | 5.07 |
ENSMUST00000156873.8
|
Chil1
|
chitinase-like 1 |
| chr1_-_13061333 | 4.87 |
ENSMUST00000115403.9
ENSMUST00000136197.8 ENSMUST00000115402.8 |
Slco5a1
|
solute carrier organic anion transporter family, member 5A1 |
| chrX_-_9983836 | 4.61 |
ENSMUST00000115543.3
ENSMUST00000044789.10 ENSMUST00000115544.9 |
Srpx
|
sushi-repeat-containing protein |
| chr1_-_128520002 | 4.47 |
ENSMUST00000052172.7
ENSMUST00000142893.2 |
Cxcr4
|
chemokine (C-X-C motif) receptor 4 |
| chr6_+_41107047 | 4.37 |
ENSMUST00000103271.2
|
Trbv13-3
|
T cell receptor beta, variable 13-3 |
| chr11_+_49554430 | 4.27 |
ENSMUST00000043873.10
ENSMUST00000076006.5 |
Scgb3a1
|
secretoglobin, family 3A, member 1 |
| chr8_+_72889073 | 3.91 |
ENSMUST00000003575.11
|
Tpm4
|
tropomyosin 4 |
| chr11_-_106606076 | 3.91 |
ENSMUST00000080853.11
ENSMUST00000183610.8 ENSMUST00000103069.10 ENSMUST00000106796.9 |
Pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr18_+_60936910 | 3.88 |
ENSMUST00000097563.9
ENSMUST00000050487.16 ENSMUST00000167610.2 |
Cd74
|
CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated) |
| chr8_+_72889607 | 3.84 |
ENSMUST00000238492.2
|
Tpm4
|
tropomyosin 4 |
| chr1_-_72914036 | 3.59 |
ENSMUST00000027377.9
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr2_+_32766126 | 3.55 |
ENSMUST00000028135.15
|
Niban2
|
niban apoptosis regulator 2 |
| chr14_+_75373766 | 3.51 |
ENSMUST00000145303.8
|
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr7_+_24069680 | 3.43 |
ENSMUST00000205428.2
ENSMUST00000171904.3 ENSMUST00000205626.2 |
Kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr15_+_73620213 | 3.38 |
ENSMUST00000053232.8
|
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr12_-_36092475 | 3.36 |
ENSMUST00000020896.17
|
Tspan13
|
tetraspanin 13 |
| chr17_-_31363245 | 3.33 |
ENSMUST00000024826.8
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr15_+_75027089 | 3.32 |
ENSMUST00000190262.2
|
Ly6g
|
lymphocyte antigen 6 complex, locus G |
| chr11_-_106605772 | 3.29 |
ENSMUST00000124958.3
|
Pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr6_+_70699822 | 3.27 |
ENSMUST00000198234.2
ENSMUST00000103406.2 |
Igkj2
|
immunoglobulin kappa joining 2 |
| chr6_+_65648574 | 3.21 |
ENSMUST00000054351.6
|
Ndnf
|
neuron-derived neurotrophic factor |
| chr5_-_103777145 | 3.21 |
ENSMUST00000031263.2
|
Slc10a6
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 6 |
| chr12_-_115832846 | 3.21 |
ENSMUST00000199373.2
|
Ighv1-78
|
immunoglobulin heavy variable 1-78 |
| chr1_-_173703424 | 3.17 |
ENSMUST00000186442.7
|
Mndal
|
myeloid nuclear differentiation antigen like |
| chr17_+_35481702 | 3.14 |
ENSMUST00000172785.8
|
H2-D1
|
histocompatibility 2, D region locus 1 |
| chr10_+_96453408 | 3.13 |
ENSMUST00000218953.2
|
Btg1
|
BTG anti-proliferation factor 1 |
| chr10_-_37014859 | 3.08 |
ENSMUST00000092584.6
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chrX_-_165992311 | 3.06 |
ENSMUST00000112172.4
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr4_+_109272828 | 3.06 |
ENSMUST00000106618.8
|
Ttc39a
|
tetratricopeptide repeat domain 39A |
| chr17_+_35278011 | 3.05 |
ENSMUST00000007255.13
ENSMUST00000174493.8 |
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr6_+_122490534 | 2.99 |
ENSMUST00000032210.14
ENSMUST00000148517.8 |
Mfap5
|
microfibrillar associated protein 5 |
| chr17_-_18104182 | 2.98 |
ENSMUST00000061516.8
|
Fpr1
|
formyl peptide receptor 1 |
| chr4_-_130068902 | 2.93 |
ENSMUST00000105998.8
|
Tinagl1
|
tubulointerstitial nephritis antigen-like 1 |
| chr6_+_41155309 | 2.89 |
ENSMUST00000103276.3
|
Trbv19
|
T cell receptor beta, variable 19 |
| chrX_+_133486391 | 2.86 |
ENSMUST00000113211.8
|
Rpl36a
|
ribosomal protein L36A |
| chr17_-_36149142 | 2.78 |
ENSMUST00000001566.10
|
Tubb5
|
tubulin, beta 5 class I |
| chr1_+_174158083 | 2.76 |
ENSMUST00000027816.5
|
Mptx1
|
mucosal pentraxin 1 |
| chr5_-_137869969 | 2.75 |
ENSMUST00000196162.5
|
Pilrb2
|
paired immunoglobin-like type 2 receptor beta 2 |
| chr6_-_128765529 | 2.74 |
ENSMUST00000167691.9
ENSMUST00000174404.8 |
Klrb1c
|
killer cell lectin-like receptor subfamily B member 1C |
| chr18_+_89215575 | 2.66 |
ENSMUST00000097496.4
ENSMUST00000236452.2 |
Cd226
|
CD226 antigen |
| chr1_-_64161415 | 2.64 |
ENSMUST00000135075.2
|
Klf7
|
Kruppel-like factor 7 (ubiquitous) |
| chr9_+_65172455 | 2.61 |
ENSMUST00000048762.8
|
Cilp
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
| chr1_-_180641099 | 2.53 |
ENSMUST00000159789.2
ENSMUST00000081026.11 |
H3f3a
|
H3.3 histone A |
| chr17_-_36149100 | 2.53 |
ENSMUST00000134978.3
|
Tubb5
|
tubulin, beta 5 class I |
| chr1_-_180641159 | 2.51 |
ENSMUST00000162118.8
ENSMUST00000159685.2 ENSMUST00000161308.8 |
H3f3a
|
H3.3 histone A |
| chr11_+_58531220 | 2.50 |
ENSMUST00000075084.5
|
Trim58
|
tripartite motif-containing 58 |
| chr17_+_35643818 | 2.50 |
ENSMUST00000174699.8
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr1_-_155022501 | 2.49 |
ENSMUST00000027744.10
|
Mr1
|
major histocompatibility complex, class I-related |
| chr17_+_35643853 | 2.41 |
ENSMUST00000113879.4
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr2_+_59314989 | 2.40 |
ENSMUST00000028369.6
|
Dapl1
|
death associated protein-like 1 |
| chr1_-_180641430 | 2.40 |
ENSMUST00000162814.8
|
H3f3a
|
H3.3 histone A |
| chr2_+_119449192 | 2.39 |
ENSMUST00000028771.8
|
Nusap1
|
nucleolar and spindle associated protein 1 |
| chr5_+_115039359 | 2.39 |
ENSMUST00000124716.3
|
Oasl2
|
2'-5' oligoadenylate synthetase-like 2 |
| chr16_+_37597235 | 2.36 |
ENSMUST00000114763.3
|
Fstl1
|
follistatin-like 1 |
| chr11_+_116421923 | 2.33 |
ENSMUST00000138840.8
|
Sphk1
|
sphingosine kinase 1 |
| chr19_-_57227742 | 2.32 |
ENSMUST00000111559.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr7_-_100164007 | 2.32 |
ENSMUST00000207405.2
|
Dnajb13
|
DnaJ heat shock protein family (Hsp40) member B13 |
| chr9_-_32452885 | 2.31 |
ENSMUST00000016231.14
|
Fli1
|
Friend leukemia integration 1 |
| chr19_-_57107413 | 2.28 |
ENSMUST00000111528.8
ENSMUST00000111529.8 ENSMUST00000104902.9 |
Ablim1
|
actin-binding LIM protein 1 |
| chr17_-_79662514 | 2.27 |
ENSMUST00000068958.9
|
Cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr10_-_18890281 | 2.26 |
ENSMUST00000146388.2
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr3_+_89986831 | 2.18 |
ENSMUST00000029549.16
|
Tpm3
|
tropomyosin 3, gamma |
| chr9_+_95519654 | 2.17 |
ENSMUST00000015498.9
|
Pcolce2
|
procollagen C-endopeptidase enhancer 2 |
| chr13_-_22016364 | 2.10 |
ENSMUST00000102979.2
|
H4c18
|
H4 clustered histone 18 |
| chr17_-_36014892 | 2.10 |
ENSMUST00000097333.10
ENSMUST00000003628.13 |
Ddr1
|
discoidin domain receptor family, member 1 |
| chr19_-_47907705 | 2.09 |
ENSMUST00000095998.7
|
Itprip
|
inositol 1,4,5-triphosphate receptor interacting protein |
| chr18_+_89215543 | 2.06 |
ENSMUST00000037142.13
|
Cd226
|
CD226 antigen |
| chr3_+_60503051 | 2.05 |
ENSMUST00000192757.6
ENSMUST00000193518.6 ENSMUST00000195817.3 |
Mbnl1
|
muscleblind like splicing factor 1 |
| chr6_-_69753317 | 2.04 |
ENSMUST00000103366.3
|
Igkv5-45
|
immunoglobulin kappa chain variable 5-45 |
| chr3_-_37778470 | 1.99 |
ENSMUST00000108105.2
ENSMUST00000079755.5 ENSMUST00000099128.2 |
Gm5148
|
predicted gene 5148 |
| chr3_-_144525255 | 1.99 |
ENSMUST00000029929.12
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr1_+_11063678 | 1.98 |
ENSMUST00000027056.12
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr4_+_57821050 | 1.95 |
ENSMUST00000238994.2
|
Pakap
|
paralemmin A kinase anchor protein |
| chr14_+_76082533 | 1.95 |
ENSMUST00000110894.9
|
Tpt1
|
tumor protein, translationally-controlled 1 |
| chr15_+_75468473 | 1.93 |
ENSMUST00000189944.7
ENSMUST00000023243.11 |
Gpihbp1
|
GPI-anchored HDL-binding protein 1 |
| chr9_-_103357564 | 1.93 |
ENSMUST00000124310.5
|
Bfsp2
|
beaded filament structural protein 2, phakinin |
| chr6_+_70700522 | 1.92 |
ENSMUST00000103408.2
|
Igkj4
|
immunoglobulin kappa joining 4 |
| chr6_-_82751429 | 1.91 |
ENSMUST00000000642.11
|
Hk2
|
hexokinase 2 |
| chr12_+_116369017 | 1.90 |
ENSMUST00000084828.5
ENSMUST00000222469.2 ENSMUST00000221114.2 ENSMUST00000221970.2 |
Ncapg2
|
non-SMC condensin II complex, subunit G2 |
| chr12_+_108572015 | 1.89 |
ENSMUST00000109854.9
|
Evl
|
Ena-vasodilator stimulated phosphoprotein |
| chr10_+_126899468 | 1.88 |
ENSMUST00000120226.8
ENSMUST00000133115.8 |
Cdk4
|
cyclin-dependent kinase 4 |
| chr16_-_18405709 | 1.87 |
ENSMUST00000232335.2
|
Tbx1
|
T-box 1 |
| chr8_+_95537074 | 1.87 |
ENSMUST00000034232.3
|
Ccl17
|
chemokine (C-C motif) ligand 17 |
| chr1_+_135768595 | 1.85 |
ENSMUST00000112087.9
ENSMUST00000178854.8 ENSMUST00000027671.12 ENSMUST00000179863.8 ENSMUST00000112085.9 ENSMUST00000112086.3 |
Tnnt2
|
troponin T2, cardiac |
| chr14_+_76082736 | 1.85 |
ENSMUST00000142061.3
|
Tpt1
|
tumor protein, translationally-controlled 1 |
| chr9_+_122863791 | 1.84 |
ENSMUST00000026893.6
ENSMUST00000215247.2 |
Tgm4
|
transglutaminase 4 (prostate) |
| chr3_-_83947416 | 1.81 |
ENSMUST00000192095.6
ENSMUST00000191758.6 ENSMUST00000052342.9 |
Tmem131l
|
transmembrane 131 like |
| chr2_+_120307390 | 1.78 |
ENSMUST00000110716.9
ENSMUST00000028748.14 ENSMUST00000090028.13 ENSMUST00000110719.4 |
Capn3
|
calpain 3 |
| chr3_+_122523219 | 1.77 |
ENSMUST00000200389.2
|
Pde5a
|
phosphodiesterase 5A, cGMP-specific |
| chr3_+_89986925 | 1.76 |
ENSMUST00000118566.8
ENSMUST00000119158.8 |
Tpm3
|
tropomyosin 3, gamma |
| chr2_+_103242027 | 1.76 |
ENSMUST00000239273.2
ENSMUST00000164172.8 |
Elf5
|
E74-like factor 5 |
| chr5_+_33261563 | 1.75 |
ENSMUST00000011178.5
|
Slc5a1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr4_+_101504909 | 1.74 |
ENSMUST00000030254.15
|
Leprot
|
leptin receptor overlapping transcript |
| chr2_-_60793536 | 1.74 |
ENSMUST00000028347.13
|
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr4_+_101504938 | 1.73 |
ENSMUST00000106927.2
|
Leprot
|
leptin receptor overlapping transcript |
| chr5_-_33432310 | 1.72 |
ENSMUST00000201372.3
ENSMUST00000202962.4 ENSMUST00000201575.4 ENSMUST00000202868.4 ENSMUST00000079746.10 |
Ctbp1
|
C-terminal binding protein 1 |
| chr6_+_68547717 | 1.71 |
ENSMUST00000196839.5
ENSMUST00000103327.3 |
Igkv12-98
|
immunoglobulin kappa variable 12-98 |
| chr1_-_173105344 | 1.68 |
ENSMUST00000111224.5
|
Mptx2
|
mucosal pentraxin 2 |
| chr15_-_12549963 | 1.68 |
ENSMUST00000189324.2
|
Pdzd2
|
PDZ domain containing 2 |
| chr16_-_56616186 | 1.67 |
ENSMUST00000023437.5
|
Adgrg7
|
adhesion G protein-coupled receptor G7 |
| chr6_-_128765449 | 1.66 |
ENSMUST00000172601.8
|
Klrb1c
|
killer cell lectin-like receptor subfamily B member 1C |
| chr14_+_70768257 | 1.66 |
ENSMUST00000047331.8
|
Lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr1_+_39940189 | 1.66 |
ENSMUST00000191761.6
ENSMUST00000193682.6 ENSMUST00000195860.6 ENSMUST00000195259.6 ENSMUST00000195636.6 ENSMUST00000192509.6 |
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr2_-_179618439 | 1.66 |
ENSMUST00000041618.13
ENSMUST00000227325.2 |
Taf4
|
TATA-box binding protein associated factor 4 |
| chr11_-_97913420 | 1.65 |
ENSMUST00000103144.10
ENSMUST00000017552.13 ENSMUST00000092736.11 ENSMUST00000107562.2 |
Cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr14_+_53698556 | 1.64 |
ENSMUST00000181728.3
|
Trav7-4
|
T cell receptor alpha variable 7-4 |
| chr14_-_51295099 | 1.63 |
ENSMUST00000227764.2
|
Rnase12
|
ribonuclease, RNase A family, 12 (non-active) |
| chrX_-_100777806 | 1.63 |
ENSMUST00000056614.7
|
Cxcr3
|
chemokine (C-X-C motif) receptor 3 |
| chr12_-_32111214 | 1.59 |
ENSMUST00000003079.12
ENSMUST00000036497.16 |
Prkar2b
|
protein kinase, cAMP dependent regulatory, type II beta |
| chr12_+_18564466 | 1.58 |
ENSMUST00000177778.9
ENSMUST00000063216.6 |
5730507C01Rik
|
RIKEN cDNA 5730507C01 gene |
| chr4_+_122910382 | 1.57 |
ENSMUST00000102649.4
|
Trit1
|
tRNA isopentenyltransferase 1 |
| chr17_-_36015484 | 1.56 |
ENSMUST00000117301.8
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr1_-_16174387 | 1.55 |
ENSMUST00000149566.2
|
Rpl7
|
ribosomal protein L7 |
| chr1_+_90131702 | 1.54 |
ENSMUST00000065587.5
ENSMUST00000159654.2 |
Ackr3
|
atypical chemokine receptor 3 |
| chr12_+_35975619 | 1.53 |
ENSMUST00000042101.5
ENSMUST00000154042.2 |
Agr3
|
anterior gradient 3 |
| chr1_-_171854818 | 1.53 |
ENSMUST00000138714.2
ENSMUST00000027837.13 ENSMUST00000111264.8 |
Vangl2
|
VANGL planar cell polarity 2 |
| chr9_+_123901979 | 1.52 |
ENSMUST00000171719.8
|
Ccr2
|
chemokine (C-C motif) receptor 2 |
| chr10_-_117212826 | 1.52 |
ENSMUST00000177145.8
ENSMUST00000176670.8 |
Cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr3_-_103553347 | 1.49 |
ENSMUST00000117271.3
|
Atg4a-ps
|
autophagy related 4A, pseudogene |
| chr15_+_75140241 | 1.47 |
ENSMUST00000023247.13
|
Ly6f
|
lymphocyte antigen 6 complex, locus F |
| chrX_+_76554608 | 1.45 |
ENSMUST00000088217.12
|
Tbl1x
|
transducin (beta)-like 1 X-linked |
| chr2_-_11506511 | 1.44 |
ENSMUST00000183869.8
|
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr7_+_121818692 | 1.42 |
ENSMUST00000033152.5
|
Chp2
|
calcineurin-like EF hand protein 2 |
| chr6_+_70699533 | 1.42 |
ENSMUST00000103405.2
|
Igkj1
|
immunoglobulin kappa joining 1 |
| chr10_-_91007387 | 1.39 |
ENSMUST00000099355.12
ENSMUST00000105293.11 ENSMUST00000092219.14 ENSMUST00000020123.7 ENSMUST00000072239.14 |
Tmpo
|
thymopoietin |
| chr13_-_89890363 | 1.38 |
ENSMUST00000109543.9
ENSMUST00000159337.8 ENSMUST00000159910.8 ENSMUST00000109544.9 |
Vcan
|
versican |
| chr11_+_67128843 | 1.38 |
ENSMUST00000018632.11
|
Myh4
|
myosin, heavy polypeptide 4, skeletal muscle |
| chr11_+_60590498 | 1.36 |
ENSMUST00000052346.10
|
Llgl1
|
LLGL1 scribble cell polarity complex component |
| chr8_-_78451055 | 1.36 |
ENSMUST00000034029.8
|
Ednra
|
endothelin receptor type A |
| chr4_-_136329953 | 1.36 |
ENSMUST00000105847.8
ENSMUST00000116273.9 |
Kdm1a
|
lysine (K)-specific demethylase 1A |
| chr1_+_177270101 | 1.35 |
ENSMUST00000194319.2
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr13_+_21995906 | 1.35 |
ENSMUST00000104941.4
|
H4c17
|
H4 clustered histone 17 |
| chr4_+_137944546 | 1.34 |
ENSMUST00000130071.2
|
Hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr2_-_7086066 | 1.34 |
ENSMUST00000183209.8
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chrX_-_135893776 | 1.34 |
ENSMUST00000171738.2
ENSMUST00000056674.6 ENSMUST00000129807.2 |
Slc25a53
|
solute carrier family 25, member 53 |
| chr9_-_88364593 | 1.33 |
ENSMUST00000173801.8
ENSMUST00000069221.12 ENSMUST00000172508.2 |
Syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr5_-_109207435 | 1.31 |
ENSMUST00000164875.3
|
Vmn2r11
|
vomeronasal 2, receptor 11 |
| chr15_-_96540760 | 1.29 |
ENSMUST00000088452.11
|
Slc38a1
|
solute carrier family 38, member 1 |
| chr11_-_79145489 | 1.29 |
ENSMUST00000017821.12
|
Wsb1
|
WD repeat and SOCS box-containing 1 |
| chr8_+_106785434 | 1.28 |
ENSMUST00000212742.2
ENSMUST00000211991.2 |
Nfatc3
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 3 |
| chr9_-_66032134 | 1.26 |
ENSMUST00000034946.15
|
Snx1
|
sorting nexin 1 |
| chr17_+_48623157 | 1.26 |
ENSMUST00000049614.13
|
B430306N03Rik
|
RIKEN cDNA B430306N03 gene |
| chr3_+_89622323 | 1.24 |
ENSMUST00000098924.9
|
Adar
|
adenosine deaminase, RNA-specific |
| chr17_-_48474356 | 1.22 |
ENSMUST00000027764.10
ENSMUST00000053612.14 |
A530064D06Rik
|
RIKEN cDNA A530064D06 gene |
| chr2_-_24809583 | 1.22 |
ENSMUST00000046227.12
ENSMUST00000114432.9 ENSMUST00000091348.11 ENSMUST00000102938.10 ENSMUST00000150379.2 ENSMUST00000152161.8 ENSMUST00000147147.8 |
Ehmt1
|
euchromatic histone methyltransferase 1 |
| chr8_+_70275079 | 1.20 |
ENSMUST00000164890.8
ENSMUST00000034325.6 ENSMUST00000238452.2 |
Lpar2
|
lysophosphatidic acid receptor 2 |
| chr19_+_44282113 | 1.20 |
ENSMUST00000026221.7
|
Scd2
|
stearoyl-Coenzyme A desaturase 2 |
| chr3_+_129695068 | 1.19 |
ENSMUST00000029626.9
|
Casp6
|
caspase 6 |
| chr12_+_105651643 | 1.18 |
ENSMUST00000051934.7
|
Gskip
|
GSK3B interacting protein |
| chr5_+_53748323 | 1.17 |
ENSMUST00000201883.4
|
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr12_-_55033130 | 1.17 |
ENSMUST00000173433.8
ENSMUST00000173803.2 |
Baz1a
Gm20403
|
bromodomain adjacent to zinc finger domain 1A predicted gene 20403 |
| chr9_-_21671571 | 1.17 |
ENSMUST00000217382.2
ENSMUST00000214149.2 ENSMUST00000098942.6 ENSMUST00000216057.2 |
Spc24
|
SPC24, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr2_-_119448935 | 1.16 |
ENSMUST00000123818.2
|
Oip5
|
Opa interacting protein 5 |
| chr12_-_55033113 | 1.16 |
ENSMUST00000038926.13
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr2_-_7086018 | 1.16 |
ENSMUST00000114923.3
ENSMUST00000182706.8 |
Celf2
|
CUGBP, Elav-like family member 2 |
| chr17_-_36014863 | 1.16 |
ENSMUST00000148065.8
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr17_-_29226886 | 1.15 |
ENSMUST00000232723.2
|
Stk38
|
serine/threonine kinase 38 |
| chr10_-_117212860 | 1.15 |
ENSMUST00000069168.13
ENSMUST00000176686.8 |
Cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr9_+_61280501 | 1.14 |
ENSMUST00000162583.8
ENSMUST00000161993.8 ENSMUST00000160882.8 ENSMUST00000160724.8 ENSMUST00000162973.8 ENSMUST00000159050.8 |
Tle3
|
transducin-like enhancer of split 3 |
| chr15_+_81469538 | 1.13 |
ENSMUST00000068387.11
|
Ep300
|
E1A binding protein p300 |
| chr7_+_99184858 | 1.12 |
ENSMUST00000032995.15
ENSMUST00000162404.8 |
Arrb1
|
arrestin, beta 1 |
| chrX_+_100473161 | 1.11 |
ENSMUST00000033673.7
|
Nono
|
non-POU-domain-containing, octamer binding protein |
| chr8_-_71349927 | 1.10 |
ENSMUST00000212709.2
ENSMUST00000212796.2 ENSMUST00000212378.2 ENSMUST00000054220.10 ENSMUST00000212494.2 |
Rpl18a
|
ribosomal protein L18A |
| chr17_-_34834938 | 1.09 |
ENSMUST00000097345.10
ENSMUST00000015611.14 |
Egfl8
|
EGF-like domain 8 |
| chr14_+_70768289 | 1.09 |
ENSMUST00000226548.2
|
Lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr15_+_88635852 | 1.07 |
ENSMUST00000041297.15
|
Zbed4
|
zinc finger, BED type containing 4 |
| chr3_-_104960264 | 1.06 |
ENSMUST00000098763.7
ENSMUST00000197437.5 |
Cttnbp2nl
|
CTTNBP2 N-terminal like |
| chr2_-_163486998 | 1.06 |
ENSMUST00000017851.4
|
Serinc3
|
serine incorporator 3 |
| chr11_-_78056347 | 1.05 |
ENSMUST00000017530.4
|
Traf4
|
TNF receptor associated factor 4 |
| chr13_+_112937385 | 1.05 |
ENSMUST00000070951.8
|
Plpp1
|
phospholipid phosphatase 1 |
| chr3_+_7568481 | 1.02 |
ENSMUST00000051064.9
ENSMUST00000193010.2 |
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr4_+_137944438 | 1.00 |
ENSMUST00000137851.8
ENSMUST00000165861.8 |
Hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr17_-_36015496 | 0.99 |
ENSMUST00000134995.8
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr14_+_26414422 | 0.97 |
ENSMUST00000022433.12
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr3_+_146205864 | 0.97 |
ENSMUST00000119130.2
|
Gng5
|
guanine nucleotide binding protein (G protein), gamma 5 |
| chr14_+_54082691 | 0.95 |
ENSMUST00000103674.6
|
Trav19
|
T cell receptor alpha variable 19 |
| chr7_+_79675727 | 0.95 |
ENSMUST00000049680.10
|
Zfp710
|
zinc finger protein 710 |
| chr4_+_126915104 | 0.95 |
ENSMUST00000030623.8
|
Sfpq
|
splicing factor proline/glutamine rich (polypyrimidine tract binding protein associated) |
| chr14_+_30856687 | 0.94 |
ENSMUST00000090212.5
|
Nt5dc2
|
5'-nucleotidase domain containing 2 |
| chr11_-_3359162 | 0.94 |
ENSMUST00000101638.4
|
Limk2
|
LIM motif-containing protein kinase 2 |
| chr13_+_44994167 | 0.93 |
ENSMUST00000173906.3
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr5_+_21850975 | 0.93 |
ENSMUST00000095495.3
|
Armc10
|
armadillo repeat containing 10 |
| chr9_-_82856321 | 0.92 |
ENSMUST00000189985.2
|
Phip
|
pleckstrin homology domain interacting protein |
| chr14_+_53007210 | 0.91 |
ENSMUST00000178768.4
|
Trav7d-4
|
T cell receptor alpha variable 7D-4 |
| chr10_+_44943262 | 0.90 |
ENSMUST00000099858.4
|
Prep
|
prolyl endopeptidase |
| chr15_-_76809607 | 0.90 |
ENSMUST00000229865.2
ENSMUST00000229055.2 |
Zfp647
|
zinc finger protein 647 |
| chr9_+_61280764 | 0.89 |
ENSMUST00000160541.8
ENSMUST00000161207.8 ENSMUST00000159630.8 |
Tle3
|
transducin-like enhancer of split 3 |
| chr17_+_14087827 | 0.89 |
ENSMUST00000239324.2
|
Afdn
|
afadin, adherens junction formation factor |
| chr2_+_145627900 | 0.89 |
ENSMUST00000110005.8
ENSMUST00000094480.11 |
Rin2
|
Ras and Rab interactor 2 |
| chr19_+_6413840 | 0.89 |
ENSMUST00000113488.8
ENSMUST00000113487.8 |
Sf1
|
splicing factor 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.2 | GO:0090673 | endothelial cell-matrix adhesion(GO:0090673) |
| 2.3 | 7.0 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 2.2 | 6.6 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 1.8 | 5.4 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 1.5 | 7.4 | GO:0031509 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 1.3 | 5.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.9 | 3.8 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.9 | 1.8 | GO:0042628 | mating plug formation(GO:0042628) single-organism reproductive behavior(GO:0044704) post-mating behavior(GO:0045297) |
| 0.9 | 3.6 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.9 | 4.5 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.9 | 2.6 | GO:0090320 | regulation of chylomicron remnant clearance(GO:0090320) |
| 0.8 | 4.7 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.8 | 3.9 | GO:0002906 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.8 | 4.6 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.8 | 2.3 | GO:0034148 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.6 | 3.1 | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.6 | 1.9 | GO:0021644 | vagus nerve morphogenesis(GO:0021644) |
| 0.6 | 9.1 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.6 | 2.3 | GO:0097298 | regulation of nucleus size(GO:0097298) |
| 0.6 | 2.3 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.6 | 3.5 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.6 | 1.7 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.6 | 6.2 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.6 | 2.2 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.5 | 3.2 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.5 | 11.9 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.5 | 1.5 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.5 | 1.5 | GO:0090265 | immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) negative regulation of eosinophil activation(GO:1902567) positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) |
| 0.5 | 5.8 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.5 | 3.3 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.4 | 1.8 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.4 | 3.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.4 | 1.2 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.4 | 1.9 | GO:0035795 | negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.4 | 1.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) regulation of muscle atrophy(GO:0014735) positive regulation of muscle atrophy(GO:0014737) |
| 0.3 | 1.7 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.3 | 3.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.3 | 1.4 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.3 | 1.3 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.3 | 2.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.3 | 2.6 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.3 | 1.9 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.3 | 1.8 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.3 | 1.8 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.3 | 0.9 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.3 | 1.9 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.3 | 1.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.3 | 1.8 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.2 | 6.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 1.2 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.2 | 3.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.2 | 1.2 | GO:1905068 | positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.2 | 3.4 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.2 | 3.4 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.2 | 1.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.2 | 1.6 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.2 | 1.2 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.2 | 1.2 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.2 | 0.7 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.2 | 1.3 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.2 | 0.9 | GO:0040010 | positive regulation of growth rate(GO:0040010) |
| 0.2 | 2.3 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.9 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.4 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 0.7 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.1 | 1.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.7 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.1 | 6.9 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 0.8 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 0.5 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 4.3 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 1.9 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 0.5 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.1 | 2.5 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 0.6 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 2.4 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 4.4 | GO:0045954 | positive regulation of natural killer cell mediated immunity(GO:0002717) positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.1 | 1.9 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 1.6 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) |
| 0.1 | 1.4 | GO:0014824 | artery smooth muscle contraction(GO:0014824) |
| 0.1 | 1.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 0.4 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.1 | 1.6 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.9 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 0.8 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.1 | 0.5 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 1.0 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.9 | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.1 | 0.6 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 0.4 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 | 4.4 | GO:0001562 | response to protozoan(GO:0001562) |
| 0.1 | 0.8 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 1.0 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 1.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 1.0 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.1 | 1.9 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.1 | 3.0 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 4.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.4 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 1.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.3 | GO:0009397 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.1 | 0.1 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.1 | 0.6 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 1.7 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 1.3 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.1 | 1.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.6 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 2.0 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.2 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 | 0.6 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.6 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 9.8 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 1.4 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) |
| 0.1 | 2.3 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.5 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.1 | 0.7 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.9 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 1.1 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 1.0 | GO:0046036 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.1 | 1.5 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 2.1 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.1 | 3.0 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.5 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.7 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 0.4 | GO:0021747 | cochlear nucleus development(GO:0021747) |
| 0.1 | 0.2 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 0.3 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.1 | 1.7 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.6 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.8 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.8 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 1.0 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.6 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 5.3 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.9 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.8 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 2.0 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.2 | GO:0060112 | positive regulation of luteinizing hormone secretion(GO:0033686) generation of ovulation cycle rhythm(GO:0060112) |
| 0.0 | 2.4 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.2 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.0 | 0.1 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 1.6 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 2.1 | GO:0006378 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 0.3 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.4 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.1 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 2.4 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.7 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.7 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.0 | 0.7 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 1.7 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.0 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 2.7 | GO:1903169 | regulation of calcium ion transmembrane transport(GO:1903169) |
| 0.0 | 1.4 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.0 | 0.1 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 5.4 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 0.5 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 0.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.6 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 0.4 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.5 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.4 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 4.1 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.8 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.0 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 1.1 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 0.5 | GO:0034766 | negative regulation of ion transmembrane transport(GO:0034766) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 6.5 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 2.3 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.5 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 1.4 | GO:0072163 | ureteric bud development(GO:0001657) mesonephric epithelium development(GO:0072163) mesonephric tubule development(GO:0072164) |
| 0.0 | 0.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.2 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 2.1 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.2 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 0.7 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 | 0.6 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 1.1 | GO:0042594 | response to starvation(GO:0042594) |
| 0.0 | 2.5 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 1.8 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.3 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.4 | GO:0001740 | Barr body(GO:0001740) |
| 1.0 | 9.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.9 | 5.4 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.9 | 10.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.8 | 7.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.8 | 11.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.8 | 10.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.6 | 3.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.6 | 2.3 | GO:0008623 | CHRAC(GO:0008623) |
| 0.5 | 1.5 | GO:0060187 | cell pole(GO:0060187) |
| 0.5 | 7.0 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.5 | 4.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.4 | 1.3 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.4 | 3.6 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.3 | 2.2 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.3 | 1.3 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.2 | 1.0 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.2 | 1.9 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.2 | 1.3 | GO:0097452 | CRD-mediated mRNA stability complex(GO:0070937) GAIT complex(GO:0097452) |
| 0.2 | 1.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.2 | 1.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.2 | 1.9 | GO:0000796 | condensin complex(GO:0000796) |
| 0.2 | 3.0 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 0.8 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 1.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.8 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 5.2 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 3.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 1.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.8 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 1.4 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 1.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 9.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 2.3 | GO:0044815 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 0.1 | 0.7 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 1.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 1.7 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 1.0 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 5.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 2.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.3 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 3.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 1.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 1.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.6 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.6 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 1.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 19.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.5 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 3.7 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.4 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.7 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 2.9 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 1.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 5.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.6 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 7.5 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 6.9 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.1 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.3 | GO:0031430 | M band(GO:0031430) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.9 | 10.8 | GO:0008061 | chitin binding(GO:0008061) |
| 0.7 | 3.0 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.6 | 3.9 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.6 | 4.5 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.5 | 1.6 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.5 | 5.8 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.5 | 2.6 | GO:0035478 | chylomicron binding(GO:0035478) |
| 0.5 | 1.5 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.5 | 8.0 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.5 | 1.4 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.5 | 3.2 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.4 | 1.2 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.4 | 2.0 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.4 | 8.1 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.4 | 3.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.4 | 3.6 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.3 | 2.2 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.3 | 1.3 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.3 | 2.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.3 | 3.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.3 | 6.6 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.3 | 0.8 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.2 | 5.4 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.2 | 1.9 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 1.0 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.2 | 3.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 1.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 2.1 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.2 | 1.2 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.2 | 2.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.2 | 13.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 0.4 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.2 | 0.6 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.2 | 1.3 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.2 | 1.7 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.2 | 0.5 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.2 | 7.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 1.7 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.2 | 8.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 1.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 5.2 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.2 | 0.6 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.2 | 1.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.2 | 1.3 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.2 | 0.5 | GO:0016419 | [acyl-carrier-protein] S-malonyltransferase activity(GO:0004314) oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) S-acetyltransferase activity(GO:0016418) S-malonyltransferase activity(GO:0016419) malonyltransferase activity(GO:0016420) phosphopantetheine binding(GO:0031177) |
| 0.2 | 2.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 1.9 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 1.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 1.7 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.6 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 3.8 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 2.4 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 1.1 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 0.6 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 1.6 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 1.6 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 1.5 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 2.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 2.8 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.3 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.1 | 0.3 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.1 | 1.2 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 10.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 2.3 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 1.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 15.5 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 0.5 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 1.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 9.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.9 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 0.2 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.1 | 0.3 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.1 | 0.5 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.3 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 3.0 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 3.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 1.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 1.8 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.1 | 0.7 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.9 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.2 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.9 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 2.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.8 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 2.5 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 1.8 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 1.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.6 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 1.9 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.6 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 11.7 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 1.9 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 1.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 1.0 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 1.9 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 4.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 3.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 3.7 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.9 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 4.3 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.4 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.2 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 1.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 4.4 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 1.1 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 11.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 5.2 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 1.3 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 1.6 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 3.4 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.0 | GO:0032090 | Pyrin domain binding(GO:0032090) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 10.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 3.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 7.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 9.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 3.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 4.5 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 25.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 5.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 3.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 2.9 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 5.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 2.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 4.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 1.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 2.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.6 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.2 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.6 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.4 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.3 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 1.2 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 1.1 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.0 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 7.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.0 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 2.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 2.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.9 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.5 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.5 | 7.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.3 | 7.8 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.2 | 3.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 11.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 4.6 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 1.5 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 2.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 3.6 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 1.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 4.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 3.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.9 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 8.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 2.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 5.3 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.1 | 2.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 2.6 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 1.9 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 1.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 2.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 0.8 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.1 | 1.6 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 2.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 5.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 2.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.8 | REACTOME G PROTEIN BETA GAMMA SIGNALLING | Genes involved in G-protein beta:gamma signalling |
| 0.0 | 1.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.8 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 1.9 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 2.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 3.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 2.9 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |