Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
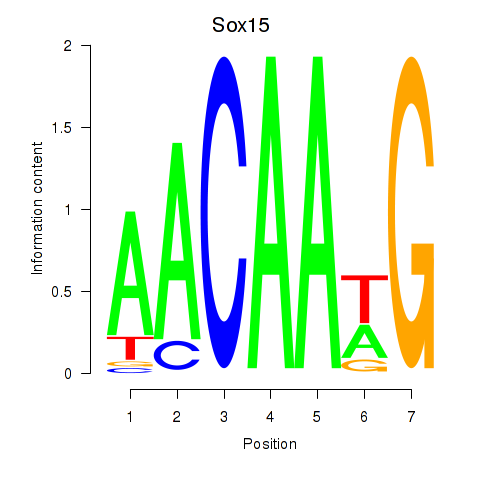
Results for Sox15
Z-value: 0.57
Transcription factors associated with Sox15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox15
|
ENSMUSG00000041287.6 | Sox15 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox15 | mm39_v1_chr11_+_69546140_69546140 | -0.27 | 2.2e-02 | Click! |
Activity profile of Sox15 motif
Sorted Z-values of Sox15 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox15
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_134944366 | 5.08 |
ENSMUST00000008987.5
|
Cldn13
|
claudin 13 |
| chr11_+_11635908 | 3.71 |
ENSMUST00000065433.12
|
Ikzf1
|
IKAROS family zinc finger 1 |
| chr16_-_45940602 | 3.63 |
ENSMUST00000023336.10
|
Cd96
|
CD96 antigen |
| chr4_+_32238950 | 3.31 |
ENSMUST00000037416.13
|
Bach2
|
BTB and CNC homology, basic leucine zipper transcription factor 2 |
| chr14_-_71003973 | 2.69 |
ENSMUST00000226448.2
ENSMUST00000022696.8 |
Xpo7
|
exportin 7 |
| chr14_-_71004019 | 2.60 |
ENSMUST00000167242.8
|
Xpo7
|
exportin 7 |
| chr7_+_121818692 | 2.26 |
ENSMUST00000033152.5
|
Chp2
|
calcineurin-like EF hand protein 2 |
| chr12_+_24758724 | 2.26 |
ENSMUST00000153058.8
|
Rrm2
|
ribonucleotide reductase M2 |
| chr12_+_24758968 | 2.09 |
ENSMUST00000154588.2
|
Rrm2
|
ribonucleotide reductase M2 |
| chr10_-_37014859 | 2.02 |
ENSMUST00000092584.6
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr10_+_87695352 | 1.93 |
ENSMUST00000121952.8
ENSMUST00000126490.8 |
Igf1
|
insulin-like growth factor 1 |
| chr9_-_103357564 | 1.82 |
ENSMUST00000124310.5
|
Bfsp2
|
beaded filament structural protein 2, phakinin |
| chr14_-_122202599 | 1.79 |
ENSMUST00000049872.9
|
Gpr183
|
G protein-coupled receptor 183 |
| chr19_-_7218363 | 1.79 |
ENSMUST00000236769.2
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr19_-_7218512 | 1.66 |
ENSMUST00000025675.11
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr17_-_71158052 | 1.66 |
ENSMUST00000186358.6
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr19_-_41836514 | 1.65 |
ENSMUST00000059231.4
|
Frat2
|
frequently rearranged in advanced T cell lymphomas 2 |
| chr2_+_146063841 | 1.63 |
ENSMUST00000089257.6
|
Insm1
|
insulinoma-associated 1 |
| chr10_+_87695117 | 1.62 |
ENSMUST00000105300.9
|
Igf1
|
insulin-like growth factor 1 |
| chr5_+_65505657 | 1.57 |
ENSMUST00000031096.11
|
Klb
|
klotho beta |
| chr18_+_11766333 | 1.50 |
ENSMUST00000115861.9
|
Rbbp8
|
retinoblastoma binding protein 8, endonuclease |
| chr10_+_87926932 | 1.49 |
ENSMUST00000048621.8
|
Pmch
|
pro-melanin-concentrating hormone |
| chr2_-_6726998 | 1.49 |
ENSMUST00000182657.2
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr10_+_79986280 | 1.47 |
ENSMUST00000153477.8
|
Midn
|
midnolin |
| chrX_+_70600481 | 1.45 |
ENSMUST00000123100.2
|
Hmgb3
|
high mobility group box 3 |
| chr11_-_106240215 | 1.44 |
ENSMUST00000021056.8
|
Scn4a
|
sodium channel, voltage-gated, type IV, alpha |
| chr9_+_96140781 | 1.41 |
ENSMUST00000190104.7
ENSMUST00000179416.8 ENSMUST00000189606.7 |
Tfdp2
|
transcription factor Dp 2 |
| chr7_-_49286594 | 1.38 |
ENSMUST00000032717.7
|
Dbx1
|
developing brain homeobox 1 |
| chr17_-_71158184 | 1.36 |
ENSMUST00000059775.15
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr2_-_65068917 | 1.30 |
ENSMUST00000090896.10
ENSMUST00000155082.2 |
Cobll1
|
Cobl-like 1 |
| chr11_+_61544085 | 1.28 |
ENSMUST00000004959.3
|
Grap
|
GRB2-related adaptor protein |
| chr11_-_100160697 | 1.24 |
ENSMUST00000017270.8
|
Krt42
|
keratin 42 |
| chr6_-_47790272 | 1.24 |
ENSMUST00000077290.9
|
Pdia4
|
protein disulfide isomerase associated 4 |
| chr11_+_45946800 | 1.20 |
ENSMUST00000011400.8
|
Adam19
|
a disintegrin and metallopeptidase domain 19 (meltrin beta) |
| chr6_-_149003003 | 1.15 |
ENSMUST00000127727.2
|
Dennd5b
|
DENN/MADD domain containing 5B |
| chr3_+_121220146 | 1.11 |
ENSMUST00000029773.13
|
Cnn3
|
calponin 3, acidic |
| chr16_-_18440388 | 1.10 |
ENSMUST00000167388.3
|
Gp1bb
|
glycoprotein Ib, beta polypeptide |
| chr13_+_94954202 | 1.08 |
ENSMUST00000220825.2
|
Tbca
|
tubulin cofactor A |
| chr11_+_117223161 | 1.08 |
ENSMUST00000106349.2
|
Septin9
|
septin 9 |
| chr4_-_34882917 | 1.07 |
ENSMUST00000098163.9
ENSMUST00000047950.6 |
Zfp292
|
zinc finger protein 292 |
| chr1_+_39940189 | 1.04 |
ENSMUST00000191761.6
ENSMUST00000193682.6 ENSMUST00000195860.6 ENSMUST00000195259.6 ENSMUST00000195636.6 ENSMUST00000192509.6 |
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr7_-_19595221 | 1.03 |
ENSMUST00000014830.8
|
Ceacam16
|
carcinoembryonic antigen-related cell adhesion molecule 16 |
| chr6_-_148846247 | 1.02 |
ENSMUST00000111562.8
ENSMUST00000081956.12 |
Sinhcaf
|
SIN3-HDAC complex associated factor |
| chr12_-_83643883 | 1.02 |
ENSMUST00000221919.2
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr5_+_53748323 | 1.00 |
ENSMUST00000201883.4
|
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr4_+_116414855 | 1.00 |
ENSMUST00000030460.15
|
Gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr10_-_89270527 | 0.99 |
ENSMUST00000218764.2
|
Gas2l3
|
growth arrest-specific 2 like 3 |
| chr1_+_86454431 | 0.97 |
ENSMUST00000045897.15
ENSMUST00000186255.7 ENSMUST00000188699.7 |
Ptma
|
prothymosin alpha |
| chr1_+_86454511 | 0.97 |
ENSMUST00000188533.2
|
Ptma
|
prothymosin alpha |
| chr18_+_34758062 | 0.96 |
ENSMUST00000166044.3
|
Kif20a
|
kinesin family member 20A |
| chr16_+_57369595 | 0.96 |
ENSMUST00000159414.2
|
Filip1l
|
filamin A interacting protein 1-like |
| chr10_+_53472853 | 0.95 |
ENSMUST00000219271.2
|
Asf1a
|
anti-silencing function 1A histone chaperone |
| chr11_+_118913788 | 0.94 |
ENSMUST00000026662.8
|
Cbx2
|
chromobox 2 |
| chr10_-_89270554 | 0.91 |
ENSMUST00000220071.2
|
Gas2l3
|
growth arrest-specific 2 like 3 |
| chr11_-_50183129 | 0.90 |
ENSMUST00000059458.5
|
Maml1
|
mastermind like transcriptional coactivator 1 |
| chr9_+_54771064 | 0.89 |
ENSMUST00000034843.9
|
Ireb2
|
iron responsive element binding protein 2 |
| chr2_-_84652890 | 0.87 |
ENSMUST00000028471.6
|
Smtnl1
|
smoothelin-like 1 |
| chr18_+_34757666 | 0.87 |
ENSMUST00000167161.9
|
Kif20a
|
kinesin family member 20A |
| chr15_-_65784103 | 0.86 |
ENSMUST00000079776.14
|
Oc90
|
otoconin 90 |
| chr10_+_87694924 | 0.85 |
ENSMUST00000095360.11
|
Igf1
|
insulin-like growth factor 1 |
| chr15_-_65784246 | 0.83 |
ENSMUST00000060522.11
|
Oc90
|
otoconin 90 |
| chr10_+_53473032 | 0.83 |
ENSMUST00000020004.8
|
Asf1a
|
anti-silencing function 1A histone chaperone |
| chr1_-_138770167 | 0.83 |
ENSMUST00000112026.4
ENSMUST00000019374.14 |
Lhx9
|
LIM homeobox protein 9 |
| chr2_-_59955995 | 0.83 |
ENSMUST00000112550.8
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr11_+_97554192 | 0.82 |
ENSMUST00000044730.12
|
Mllt6
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 6 |
| chr17_-_24054717 | 0.81 |
ENSMUST00000059906.8
|
Prss33
|
protease, serine 33 |
| chr6_+_17463748 | 0.80 |
ENSMUST00000115443.8
|
Met
|
met proto-oncogene |
| chr14_-_122202508 | 0.79 |
ENSMUST00000227267.2
|
Gpr183
|
G protein-coupled receptor 183 |
| chr7_+_65759198 | 0.77 |
ENSMUST00000036372.8
|
Chsy1
|
chondroitin sulfate synthase 1 |
| chr6_-_99703344 | 0.75 |
ENSMUST00000008273.8
ENSMUST00000101120.11 ENSMUST00000203738.2 |
Prok2
|
prokineticin 2 |
| chr9_+_96141299 | 0.74 |
ENSMUST00000179065.8
|
Tfdp2
|
transcription factor Dp 2 |
| chr4_+_109835224 | 0.73 |
ENSMUST00000061187.4
|
Dmrta2
|
doublesex and mab-3 related transcription factor like family A2 |
| chr9_+_35179153 | 0.72 |
ENSMUST00000034543.5
|
Rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr6_+_15184630 | 0.72 |
ENSMUST00000115470.3
|
Foxp2
|
forkhead box P2 |
| chr3_+_32763313 | 0.71 |
ENSMUST00000126144.3
|
Actl6a
|
actin-like 6A |
| chr9_+_96141317 | 0.70 |
ENSMUST00000165768.4
|
Tfdp2
|
transcription factor Dp 2 |
| chr12_-_83643964 | 0.70 |
ENSMUST00000048319.6
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr8_-_87472562 | 0.69 |
ENSMUST00000045296.6
|
Siah1a
|
siah E3 ubiquitin protein ligase 1A |
| chr4_-_82939330 | 0.69 |
ENSMUST00000071708.12
|
Frem1
|
Fras1 related extracellular matrix protein 1 |
| chr5_-_123859070 | 0.68 |
ENSMUST00000031376.12
|
Zcchc8
|
zinc finger, CCHC domain containing 8 |
| chr4_+_116415251 | 0.68 |
ENSMUST00000106475.2
|
Gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr12_-_76842263 | 0.67 |
ENSMUST00000082431.6
|
Gpx2
|
glutathione peroxidase 2 |
| chr12_+_20004506 | 0.67 |
ENSMUST00000179133.2
|
Gm21863
|
predicted gene, 21863 |
| chr12_-_84265609 | 0.67 |
ENSMUST00000046266.13
ENSMUST00000220974.2 |
Mideas
|
mitotic deacetylase associated SANT domain protein |
| chr11_-_99742434 | 0.67 |
ENSMUST00000107437.2
|
Krtap4-16
|
keratin associated protein 4-16 |
| chr6_-_52141796 | 0.66 |
ENSMUST00000014848.11
|
Hoxa2
|
homeobox A2 |
| chr13_-_59930059 | 0.65 |
ENSMUST00000225581.2
|
Gm49354
|
predicted gene, 49354 |
| chr8_+_45960855 | 0.62 |
ENSMUST00000141039.8
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr11_+_67061837 | 0.62 |
ENSMUST00000170159.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chrX_-_56438380 | 0.59 |
ENSMUST00000143310.2
ENSMUST00000098470.9 ENSMUST00000114726.8 |
Rbmx
|
RNA binding motif protein, X chromosome |
| chr11_-_106606076 | 0.58 |
ENSMUST00000080853.11
ENSMUST00000183610.8 ENSMUST00000103069.10 ENSMUST00000106796.9 |
Pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr6_+_65019574 | 0.58 |
ENSMUST00000031984.9
ENSMUST00000205118.3 |
Smarcad1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr3_-_84387700 | 0.58 |
ENSMUST00000194027.2
ENSMUST00000107689.7 |
Fhdc1
|
FH2 domain containing 1 |
| chrX_-_58613428 | 0.57 |
ENSMUST00000119833.8
ENSMUST00000131319.8 |
Fgf13
|
fibroblast growth factor 13 |
| chr18_+_82932747 | 0.55 |
ENSMUST00000071233.7
|
Zfp516
|
zinc finger protein 516 |
| chr1_+_172327569 | 0.55 |
ENSMUST00000111230.8
|
Tagln2
|
transgelin 2 |
| chr2_+_160573604 | 0.55 |
ENSMUST00000174885.2
ENSMUST00000109462.8 |
Plcg1
|
phospholipase C, gamma 1 |
| chr6_+_54016543 | 0.55 |
ENSMUST00000046856.14
|
Chn2
|
chimerin 2 |
| chr5_-_123859153 | 0.54 |
ENSMUST00000196282.5
|
Zcchc8
|
zinc finger, CCHC domain containing 8 |
| chr1_-_119764729 | 0.53 |
ENSMUST00000163621.2
ENSMUST00000168303.8 |
Ptpn4
|
protein tyrosine phosphatase, non-receptor type 4 |
| chr1_+_31215482 | 0.53 |
ENSMUST00000062560.14
|
Lgsn
|
lengsin, lens protein with glutamine synthetase domain |
| chr18_+_34757687 | 0.53 |
ENSMUST00000237407.2
|
Kif20a
|
kinesin family member 20A |
| chr6_-_93889483 | 0.53 |
ENSMUST00000205116.3
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr6_+_65019793 | 0.52 |
ENSMUST00000204696.3
|
Smarcad1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr6_+_125529911 | 0.50 |
ENSMUST00000112254.8
ENSMUST00000112253.6 |
Vwf
|
Von Willebrand factor |
| chr11_-_99884818 | 0.48 |
ENSMUST00000105049.2
|
Krtap17-1
|
keratin associated protein 17-1 |
| chr10_-_80223475 | 0.47 |
ENSMUST00000105350.3
|
Mex3d
|
mex3 RNA binding family member D |
| chr4_+_19280850 | 0.47 |
ENSMUST00000102999.2
|
Cngb3
|
cyclic nucleotide gated channel beta 3 |
| chr10_+_29020055 | 0.47 |
ENSMUST00000216757.2
|
Soga3
|
SOGA family member 3 |
| chr18_-_46345661 | 0.47 |
ENSMUST00000037011.6
|
Trim36
|
tripartite motif-containing 36 |
| chr19_-_4384029 | 0.46 |
ENSMUST00000176653.2
|
Kdm2a
|
lysine (K)-specific demethylase 2A |
| chr17_+_52909721 | 0.45 |
ENSMUST00000039366.11
|
Kcnh8
|
potassium voltage-gated channel, subfamily H (eag-related), member 8 |
| chr6_+_17463819 | 0.45 |
ENSMUST00000140070.8
|
Met
|
met proto-oncogene |
| chr13_+_42205790 | 0.44 |
ENSMUST00000220525.2
|
Hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr5_-_24782465 | 0.43 |
ENSMUST00000030795.10
|
Abcf2
|
ATP-binding cassette, sub-family F (GCN20), member 2 |
| chr8_-_86281946 | 0.43 |
ENSMUST00000034138.7
|
Dnaja2
|
DnaJ heat shock protein family (Hsp40) member A2 |
| chr3_-_75864195 | 0.42 |
ENSMUST00000038563.14
ENSMUST00000167078.8 ENSMUST00000117242.8 |
Golim4
|
golgi integral membrane protein 4 |
| chr19_+_5742880 | 0.42 |
ENSMUST00000235661.2
|
Map3k11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr1_+_39940043 | 0.41 |
ENSMUST00000168431.7
ENSMUST00000163854.9 |
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr11_-_106605772 | 0.40 |
ENSMUST00000124958.3
|
Pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr14_+_26359390 | 0.39 |
ENSMUST00000112318.10
|
Arf4
|
ADP-ribosylation factor 4 |
| chrX_-_56438322 | 0.38 |
ENSMUST00000114730.8
|
Rbmx
|
RNA binding motif protein, X chromosome |
| chr2_-_84255602 | 0.38 |
ENSMUST00000074262.9
|
Calcrl
|
calcitonin receptor-like |
| chr6_+_86381318 | 0.36 |
ENSMUST00000113713.9
|
Tia1
|
cytotoxic granule-associated RNA binding protein 1 |
| chr2_-_146353911 | 0.36 |
ENSMUST00000109986.9
|
Ralgapa2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
| chr9_-_35179042 | 0.36 |
ENSMUST00000217306.2
ENSMUST00000125087.2 ENSMUST00000121564.8 ENSMUST00000063782.12 ENSMUST00000059057.14 |
Fam118b
|
family with sequence similarity 118, member B |
| chr2_-_146353819 | 0.34 |
ENSMUST00000131824.8
|
Ralgapa2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
| chr5_-_66238313 | 0.34 |
ENSMUST00000202700.4
ENSMUST00000094757.9 ENSMUST00000113724.6 |
Rbm47
|
RNA binding motif protein 47 |
| chr5_+_92285748 | 0.34 |
ENSMUST00000031355.10
ENSMUST00000202155.2 |
Uso1
|
USO1 vesicle docking factor |
| chr13_-_43632368 | 0.33 |
ENSMUST00000222651.2
|
Ranbp9
|
RAN binding protein 9 |
| chr14_-_48900192 | 0.33 |
ENSMUST00000122009.8
|
Otx2
|
orthodenticle homeobox 2 |
| chr2_-_89774457 | 0.33 |
ENSMUST00000090695.3
|
Olfr1259
|
olfactory receptor 1259 |
| chr4_+_102427247 | 0.32 |
ENSMUST00000097950.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr18_+_4921663 | 0.32 |
ENSMUST00000143254.8
|
Svil
|
supervillin |
| chr2_+_6327431 | 0.32 |
ENSMUST00000114937.8
|
Usp6nl
|
USP6 N-terminal like |
| chrX_-_58612709 | 0.31 |
ENSMUST00000124402.2
|
Fgf13
|
fibroblast growth factor 13 |
| chr12_+_37291728 | 0.31 |
ENSMUST00000160768.8
|
Agmo
|
alkylglycerol monooxygenase |
| chr9_+_43655230 | 0.30 |
ENSMUST00000034510.9
|
Nectin1
|
nectin cell adhesion molecule 1 |
| chr15_+_25622611 | 0.30 |
ENSMUST00000110457.8
ENSMUST00000137601.8 |
Myo10
|
myosin X |
| chr1_-_119765343 | 0.28 |
ENSMUST00000064091.12
|
Ptpn4
|
protein tyrosine phosphatase, non-receptor type 4 |
| chr12_-_40495753 | 0.28 |
ENSMUST00000069692.10
ENSMUST00000069637.15 |
Zfp277
|
zinc finger protein 277 |
| chr2_-_164876690 | 0.26 |
ENSMUST00000122070.2
ENSMUST00000121377.8 ENSMUST00000153905.2 ENSMUST00000040381.15 |
Ncoa5
|
nuclear receptor coactivator 5 |
| chr2_-_6726701 | 0.24 |
ENSMUST00000114927.9
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr9_-_106666329 | 0.24 |
ENSMUST00000046502.7
|
Rad54l2
|
RAD54 like 2 (S. cerevisiae) |
| chr16_-_10613991 | 0.23 |
ENSMUST00000189593.2
|
Prm2
|
protamine 2 |
| chr6_+_86381201 | 0.23 |
ENSMUST00000095754.10
ENSMUST00000095753.9 |
Tia1
|
cytotoxic granule-associated RNA binding protein 1 |
| chr6_+_65019558 | 0.22 |
ENSMUST00000204801.3
|
Smarcad1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr7_-_115423934 | 0.21 |
ENSMUST00000169129.8
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr6_+_86605146 | 0.21 |
ENSMUST00000043400.9
|
Asprv1
|
aspartic peptidase, retroviral-like 1 |
| chr11_+_93935066 | 0.19 |
ENSMUST00000103168.10
|
Spag9
|
sperm associated antigen 9 |
| chr4_-_20778847 | 0.18 |
ENSMUST00000102998.4
|
Nkain3
|
Na+/K+ transporting ATPase interacting 3 |
| chr1_+_172327812 | 0.18 |
ENSMUST00000192460.2
|
Tagln2
|
transgelin 2 |
| chr9_+_13677266 | 0.17 |
ENSMUST00000152532.8
|
Mtmr2
|
myotubularin related protein 2 |
| chr6_-_112364974 | 0.17 |
ENSMUST00000238755.2
ENSMUST00000060847.6 |
Ssu2
|
ssu-2 homolog (C. elegans) |
| chr1_+_87332638 | 0.16 |
ENSMUST00000173152.2
ENSMUST00000173663.2 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr1_+_180158035 | 0.14 |
ENSMUST00000070181.7
|
Itpkb
|
inositol 1,4,5-trisphosphate 3-kinase B |
| chr4_+_128582519 | 0.11 |
ENSMUST00000106080.8
|
Phc2
|
polyhomeotic 2 |
| chr4_+_5724305 | 0.11 |
ENSMUST00000108380.2
|
Fam110b
|
family with sequence similarity 110, member B |
| chr17_-_78991691 | 0.10 |
ENSMUST00000145480.2
|
Strn
|
striatin, calmodulin binding protein |
| chr11_+_93935156 | 0.10 |
ENSMUST00000024979.15
|
Spag9
|
sperm associated antigen 9 |
| chr14_+_79689230 | 0.09 |
ENSMUST00000100359.3
ENSMUST00000226192.2 |
Kbtbd6
|
kelch repeat and BTB (POZ) domain containing 6 |
| chr9_+_74883377 | 0.09 |
ENSMUST00000081746.7
|
Fam214a
|
family with sequence similarity 214, member A |
| chr2_-_6327884 | 0.09 |
ENSMUST00000238876.2
|
1700014B07Rik
|
RIKEN cDNA 1700014B07 gene |
| chr16_-_4831349 | 0.09 |
ENSMUST00000201077.2
ENSMUST00000202281.4 ENSMUST00000090453.9 ENSMUST00000023191.17 |
Rogdi
|
rogdi homolog |
| chr6_+_86381236 | 0.08 |
ENSMUST00000095752.9
ENSMUST00000130967.8 |
Tia1
|
cytotoxic granule-associated RNA binding protein 1 |
| chr10_+_17672004 | 0.08 |
ENSMUST00000037964.7
|
Txlnb
|
taxilin beta |
| chr11_+_93935021 | 0.08 |
ENSMUST00000075695.13
ENSMUST00000092777.11 |
Spag9
|
sperm associated antigen 9 |
| chr3_-_64194838 | 0.06 |
ENSMUST00000170244.3
|
Vmn2r3
|
vomeronasal 2, receptor 3 |
| chr3_+_31956656 | 0.06 |
ENSMUST00000119310.8
|
Kcnmb2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr3_-_127631068 | 0.05 |
ENSMUST00000051737.8
ENSMUST00000200409.5 |
Ap1ar
|
adaptor-related protein complex 1 associated regulatory protein |
| chr11_-_65679101 | 0.05 |
ENSMUST00000152096.8
ENSMUST00000046963.10 |
Map2k4
|
mitogen-activated protein kinase kinase 4 |
| chr11_+_87651359 | 0.04 |
ENSMUST00000039627.12
ENSMUST00000100644.10 |
Tspoap1
|
TSPO associated protein 1 |
| chr15_-_37008011 | 0.03 |
ENSMUST00000226671.2
|
Zfp706
|
zinc finger protein 706 |
| chr18_+_65209103 | 0.03 |
ENSMUST00000236764.2
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr5_+_20112704 | 0.03 |
ENSMUST00000115267.7
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr10_+_52267702 | 0.02 |
ENSMUST00000067085.7
|
Nepn
|
nephrocan |
| chr8_-_23143422 | 0.00 |
ENSMUST00000033938.7
|
Polb
|
polymerase (DNA directed), beta |
| chr12_+_3415143 | 0.00 |
ENSMUST00000020999.7
|
Kif3c
|
kinesin family member 3C |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0045660 | positive regulation of neutrophil differentiation(GO:0045660) |
| 0.9 | 2.6 | GO:0036145 | dendritic cell homeostasis(GO:0036145) |
| 0.6 | 4.4 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.5 | 3.6 | GO:0002727 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.5 | 1.5 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.4 | 1.2 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.4 | 1.6 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.3 | 1.0 | GO:0090673 | endothelial cell-matrix adhesion(GO:0090673) |
| 0.3 | 0.9 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.3 | 1.5 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.3 | 0.8 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.2 | 0.9 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.2 | 1.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.2 | 1.0 | GO:1905068 | regulation of cell adhesion involved in heart morphogenesis(GO:0061344) positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.2 | 1.6 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.2 | 4.3 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.2 | 1.5 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.2 | 1.4 | GO:0015871 | choline transport(GO:0015871) |
| 0.2 | 0.5 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.2 | 0.7 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.2 | 1.5 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.2 | 0.7 | GO:0021658 | rhombomere 3 morphogenesis(GO:0021658) |
| 0.2 | 1.8 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.7 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) epicardium-derived cardiac vascular smooth muscle cell differentiation(GO:0060983) |
| 0.1 | 3.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.9 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.1 | 0.9 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 2.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 2.3 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.8 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 0.7 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.1 | 1.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.3 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 2.4 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.1 | 1.8 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 0.4 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 1.3 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 0.8 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.5 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.7 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 1.4 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 1.9 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 0.4 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.1 | 1.4 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 2.0 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 1.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.7 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 0.4 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.5 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.1 | 5.3 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 0.7 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 | 2.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.3 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 1.7 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.5 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.0 | 1.5 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.7 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.7 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.7 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.2 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.3 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.0 | 0.4 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.3 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.2 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.6 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 1.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.4 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.6 | GO:0009409 | response to cold(GO:0009409) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.3 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.4 | 4.4 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.3 | 2.0 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.3 | 1.7 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.2 | 1.9 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.2 | 3.7 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.2 | 1.0 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 1.0 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.7 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 1.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.6 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.7 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 5.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 5.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.0 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 3.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.7 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 2.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 3.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 3.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.9 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 2.8 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.3 | GO:0043034 | costamere(GO:0043034) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.3 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.4 | 2.6 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.4 | 5.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.3 | 3.5 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.3 | 0.9 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.3 | 0.8 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.2 | 0.9 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.2 | 3.7 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.2 | 1.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 0.5 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 0.1 | 0.4 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.1 | 3.0 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 1.5 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.8 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 3.7 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.1 | 4.4 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.5 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 0.5 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.5 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.4 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 1.2 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.1 | 0.5 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 1.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.7 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.7 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 1.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 1.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.7 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 1.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 2.0 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 1.7 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 1.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.3 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.9 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.5 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 2.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 2.0 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.0 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 4.7 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 1.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 5.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 3.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 2.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 3.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 3.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.4 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 2.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.9 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 1.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 2.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 3.0 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 1.5 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 3.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.6 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 1.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.6 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 1.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.8 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |