Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Sox17
Z-value: 0.88
Transcription factors associated with Sox17
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox17
|
ENSMUSG00000025902.14 | Sox17 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox17 | mm39_v1_chr1_-_4563821_4563958 | -0.04 | 7.6e-01 | Click! |
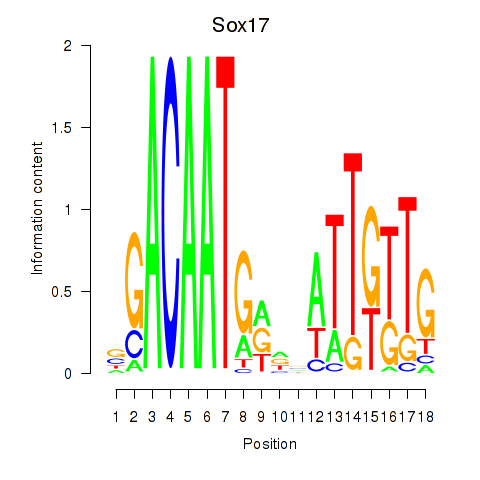
Activity profile of Sox17 motif
Sorted Z-values of Sox17 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox17
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_128503666 | 11.84 |
ENSMUST00000143664.2
ENSMUST00000112132.8 |
Pzp
|
PZP, alpha-2-macroglobulin like |
| chr16_+_22769822 | 8.96 |
ENSMUST00000023590.9
|
Hrg
|
histidine-rich glycoprotein |
| chr16_+_22769844 | 8.09 |
ENSMUST00000232422.2
|
Hrg
|
histidine-rich glycoprotein |
| chr8_-_93924426 | 7.71 |
ENSMUST00000034172.8
|
Ces1d
|
carboxylesterase 1D |
| chr5_-_89605622 | 7.38 |
ENSMUST00000049209.13
|
Gc
|
vitamin D binding protein |
| chr8_-_93956143 | 6.73 |
ENSMUST00000176282.2
ENSMUST00000034173.14 |
Ces1e
|
carboxylesterase 1E |
| chr16_+_22877000 | 6.60 |
ENSMUST00000039492.14
ENSMUST00000023589.15 ENSMUST00000089902.8 |
Kng1
|
kininogen 1 |
| chr9_-_48516447 | 6.43 |
ENSMUST00000034808.12
ENSMUST00000119426.2 |
Nnmt
|
nicotinamide N-methyltransferase |
| chr1_-_162694076 | 6.31 |
ENSMUST00000046049.14
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr3_-_98670369 | 6.03 |
ENSMUST00000107019.8
ENSMUST00000107018.8 |
Hsd3b3
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 3 |
| chr16_-_22847808 | 6.02 |
ENSMUST00000115349.9
|
Kng2
|
kininogen 2 |
| chr5_+_90708962 | 5.78 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr16_-_22847760 | 5.56 |
ENSMUST00000039338.13
|
Kng2
|
kininogen 2 |
| chr8_-_94063823 | 5.24 |
ENSMUST00000044602.8
|
Ces1g
|
carboxylesterase 1G |
| chr16_-_22847829 | 5.15 |
ENSMUST00000100046.9
|
Kng2
|
kininogen 2 |
| chrX_+_59044796 | 5.13 |
ENSMUST00000033477.5
|
F9
|
coagulation factor IX |
| chr11_-_77784922 | 5.04 |
ENSMUST00000017597.5
|
Pipox
|
pipecolic acid oxidase |
| chr16_-_22848153 | 4.11 |
ENSMUST00000232459.2
|
Kng2
|
kininogen 2 |
| chr16_-_18232202 | 4.01 |
ENSMUST00000165430.8
ENSMUST00000147720.3 |
Comt
|
catechol-O-methyltransferase |
| chr11_-_109986763 | 3.79 |
ENSMUST00000046223.14
ENSMUST00000106664.10 ENSMUST00000106662.2 |
Abca8a
|
ATP-binding cassette, sub-family A (ABC1), member 8a |
| chr11_-_109986804 | 3.65 |
ENSMUST00000100287.9
|
Abca8a
|
ATP-binding cassette, sub-family A (ABC1), member 8a |
| chr13_-_35211060 | 3.65 |
ENSMUST00000170538.8
ENSMUST00000163280.8 |
Eci2
|
enoyl-Coenzyme A delta isomerase 2 |
| chr8_+_118428643 | 3.43 |
ENSMUST00000034304.9
|
Hsd17b2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr12_+_84085709 | 3.26 |
ENSMUST00000221229.2
|
Gm49366
|
predicted gene, 49366 |
| chr12_-_40249314 | 3.24 |
ENSMUST00000095760.3
|
Lsmem1
|
leucine-rich single-pass membrane protein 1 |
| chr6_+_121709891 | 3.12 |
ENSMUST00000204124.2
|
Gm7298
|
predicted gene 7298 |
| chr7_+_44984723 | 2.83 |
ENSMUST00000211327.2
|
Hrc
|
histidine rich calcium binding protein |
| chr6_+_121222865 | 2.46 |
ENSMUST00000032198.11
|
Usp18
|
ubiquitin specific peptidase 18 |
| chr8_-_106022676 | 2.28 |
ENSMUST00000057855.4
|
Exoc3l
|
exocyst complex component 3-like |
| chr8_-_41507808 | 2.20 |
ENSMUST00000093534.11
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr5_-_38659449 | 2.14 |
ENSMUST00000005238.13
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr2_+_102489558 | 1.91 |
ENSMUST00000111213.8
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr9_+_57428490 | 1.78 |
ENSMUST00000000090.8
|
Cox5a
|
cytochrome c oxidase subunit 5A |
| chr7_+_43910845 | 1.77 |
ENSMUST00000124863.4
|
Gm15517
|
predicted gene 15517 |
| chr2_+_89808124 | 1.69 |
ENSMUST00000061830.2
|
Olfr1260
|
olfactory receptor 1260 |
| chr11_-_106278892 | 1.68 |
ENSMUST00000106813.9
ENSMUST00000141146.2 |
Icam2
|
intercellular adhesion molecule 2 |
| chr5_-_38659422 | 1.66 |
ENSMUST00000147664.8
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr9_+_66853343 | 1.63 |
ENSMUST00000040917.14
ENSMUST00000127896.8 |
Rps27l
|
ribosomal protein S27-like |
| chr10_-_102864921 | 1.58 |
ENSMUST00000217946.2
ENSMUST00000219194.2 |
Alx1
|
ALX homeobox 1 |
| chr18_+_80298441 | 1.47 |
ENSMUST00000123750.8
|
Slc66a2
|
solute carrier family 66 member 2 |
| chr11_-_86698484 | 1.43 |
ENSMUST00000018569.14
|
Dhx40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr7_-_127490139 | 1.43 |
ENSMUST00000205300.2
ENSMUST00000121394.3 |
Prss53
|
protease, serine 53 |
| chr5_+_31265651 | 1.41 |
ENSMUST00000201740.4
ENSMUST00000066544.5 |
Dnajc5g
|
DnaJ heat shock protein family (Hsp40) member C5 gamma |
| chr3_-_107240989 | 1.39 |
ENSMUST00000061772.11
|
Rbm15
|
RNA binding motif protein 15 |
| chr15_-_96358612 | 1.36 |
ENSMUST00000047835.8
|
Scaf11
|
SR-related CTD-associated factor 11 |
| chr15_+_53748302 | 1.35 |
ENSMUST00000100666.3
|
Gm7489
|
predicted gene 7489 |
| chr12_-_84447702 | 1.31 |
ENSMUST00000122194.8
|
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr19_+_6356486 | 1.31 |
ENSMUST00000025681.8
|
Cdc42bpg
|
CDC42 binding protein kinase gamma (DMPK-like) |
| chr8_+_70275079 | 1.31 |
ENSMUST00000164890.8
ENSMUST00000034325.6 ENSMUST00000238452.2 |
Lpar2
|
lysophosphatidic acid receptor 2 |
| chr3_-_92050043 | 1.28 |
ENSMUST00000197811.2
ENSMUST00000029535.6 |
4930511M18Rik
Lelp1
|
RIKEN cDNA 4930511M18 gene late cornified envelope-like proline-rich 1 |
| chr19_+_34560922 | 1.23 |
ENSMUST00000102825.4
|
Ifit3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr7_+_28937859 | 1.17 |
ENSMUST00000108237.2
|
Yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr17_+_47747657 | 1.17 |
ENSMUST00000150819.3
|
AI661453
|
expressed sequence AI661453 |
| chr7_+_28937746 | 1.13 |
ENSMUST00000108238.8
ENSMUST00000032809.10 |
Yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr4_-_131664478 | 1.13 |
ENSMUST00000155990.8
|
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr2_-_76698725 | 1.09 |
ENSMUST00000149616.8
ENSMUST00000152185.8 ENSMUST00000130915.8 ENSMUST00000155365.8 ENSMUST00000128071.8 |
Ttn
|
titin |
| chr12_-_84447625 | 1.06 |
ENSMUST00000117286.2
|
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr2_+_121287444 | 1.03 |
ENSMUST00000126764.2
|
Hypk
|
huntingtin interacting protein K |
| chr11_+_11439266 | 1.02 |
ENSMUST00000109678.3
|
Spata48
|
spermatogenesis associated 48 |
| chrX_-_47966588 | 1.02 |
ENSMUST00000114928.2
|
Fsip2l
|
fibrous sheath-interacting protein 2-like |
| chr1_-_60605867 | 1.00 |
ENSMUST00000027168.12
ENSMUST00000090293.11 ENSMUST00000140485.8 |
Raph1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr6_-_144155197 | 0.99 |
ENSMUST00000038815.14
ENSMUST00000111749.8 ENSMUST00000170367.9 |
Sox5
|
SRY (sex determining region Y)-box 5 |
| chr2_-_111100733 | 0.97 |
ENSMUST00000099619.6
|
Olfr1277
|
olfactory receptor 1277 |
| chr8_+_107877252 | 0.97 |
ENSMUST00000034400.5
|
Cyb5b
|
cytochrome b5 type B |
| chr5_-_125466240 | 0.96 |
ENSMUST00000108707.3
|
Ubc
|
ubiquitin C |
| chr14_+_53220913 | 0.94 |
ENSMUST00000167409.2
|
Trav9d-4
|
T cell receptor alpha variable 9D-4 |
| chr7_-_97760653 | 0.94 |
ENSMUST00000238309.2
|
Myo7a
|
myosin VIIA |
| chr10_+_80662490 | 0.94 |
ENSMUST00000060987.15
ENSMUST00000177850.8 ENSMUST00000180036.8 ENSMUST00000179172.8 |
Oaz1
|
ornithine decarboxylase antizyme 1 |
| chr16_+_8288627 | 0.94 |
ENSMUST00000046470.16
ENSMUST00000150790.2 ENSMUST00000142899.2 |
Mettl22
|
methyltransferase like 22 |
| chr3_+_132335575 | 0.92 |
ENSMUST00000212804.2
ENSMUST00000212852.2 |
Gimd1
|
GIMAP family P-loop NTPase domain containing 1 |
| chr14_+_73379930 | 0.92 |
ENSMUST00000170370.8
ENSMUST00000164822.8 ENSMUST00000165429.8 |
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr6_-_144155167 | 0.91 |
ENSMUST00000077160.12
|
Sox5
|
SRY (sex determining region Y)-box 5 |
| chrY_-_28226857 | 0.88 |
ENSMUST00000188864.2
|
Gm20894
|
predicted gene, 20894 |
| chr11_-_68291638 | 0.87 |
ENSMUST00000108674.9
|
Ntn1
|
netrin 1 |
| chr10_-_23226684 | 0.87 |
ENSMUST00000220299.2
|
Eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chrY_-_26732144 | 0.86 |
ENSMUST00000189518.2
|
Gm20890
|
predicted gene, 20890 |
| chrY_-_27477265 | 0.86 |
ENSMUST00000188744.2
|
Gm21488
|
predicted gene, 21488 |
| chr7_+_28937898 | 0.86 |
ENSMUST00000138128.3
ENSMUST00000142519.3 |
Yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr14_+_30856687 | 0.84 |
ENSMUST00000090212.5
|
Nt5dc2
|
5'-nucleotidase domain containing 2 |
| chr18_-_3123412 | 0.83 |
ENSMUST00000165255.3
|
Vmn1r238
|
vomeronasal 1 receptor, 238 |
| chr10_-_10348058 | 0.80 |
ENSMUST00000179956.8
ENSMUST00000208717.2 ENSMUST00000172530.8 ENSMUST00000132573.2 |
Adgb
|
androglobin |
| chr9_+_50405817 | 0.79 |
ENSMUST00000114474.8
ENSMUST00000188047.2 |
Plet1
|
placenta expressed transcript 1 |
| chr4_+_3172083 | 0.78 |
ENSMUST00000105160.5
|
Vmn1r2
|
vomeronasal 1 receptor 2 |
| chr5_-_137856280 | 0.77 |
ENSMUST00000110978.7
ENSMUST00000199387.2 ENSMUST00000196195.2 |
Pilrb1
|
paired immunoglobin-like type 2 receptor beta 1 |
| chr5_-_104169696 | 0.76 |
ENSMUST00000119025.2
|
Hsd17b11
|
hydroxysteroid (17-beta) dehydrogenase 11 |
| chr9_-_49338321 | 0.76 |
ENSMUST00000034792.7
|
Ankk1
|
ankyrin repeat and kinase domain containing 1 |
| chr7_+_104937302 | 0.76 |
ENSMUST00000098155.3
|
Olfr688
|
olfactory receptor 688 |
| chr15_-_57128522 | 0.75 |
ENSMUST00000137764.2
ENSMUST00000022995.13 |
Slc22a22
|
solute carrier family 22 (organic cation transporter), member 22 |
| chr18_+_35731658 | 0.70 |
ENSMUST00000041314.17
ENSMUST00000236666.2 ENSMUST00000236020.2 ENSMUST00000235400.2 |
Paip2
|
polyadenylate-binding protein-interacting protein 2 |
| chr2_+_111158766 | 0.68 |
ENSMUST00000090326.2
|
Olfr1281
|
olfactory receptor 1281 |
| chr9_+_40098375 | 0.67 |
ENSMUST00000062229.6
|
Olfr986
|
olfactory receptor 986 |
| chr7_+_119927885 | 0.67 |
ENSMUST00000207220.2
ENSMUST00000121265.7 ENSMUST00000076272.5 |
Abca15
|
ATP-binding cassette, sub-family A (ABC1), member 15 |
| chr17_+_47747540 | 0.66 |
ENSMUST00000037701.13
|
AI661453
|
expressed sequence AI661453 |
| chr9_+_110663656 | 0.66 |
ENSMUST00000011391.12
ENSMUST00000146794.4 |
Prss45
|
protease, serine 45 |
| chr10_-_128012336 | 0.66 |
ENSMUST00000092033.4
|
Rbms2
|
RNA binding motif, single stranded interacting protein 2 |
| chr6_+_24857967 | 0.65 |
ENSMUST00000200968.4
|
Hyal5
|
hyaluronoglucosaminidase 5 |
| chr10_-_62198043 | 0.65 |
ENSMUST00000149534.2
|
Hk1
|
hexokinase 1 |
| chr14_+_54664359 | 0.65 |
ENSMUST00000010550.12
ENSMUST00000199195.3 ENSMUST00000196273.2 |
Mrpl52
|
mitochondrial ribosomal protein L52 |
| chr19_+_12394797 | 0.64 |
ENSMUST00000216506.2
ENSMUST00000214153.2 |
Olfr1441
|
olfactory receptor 1441 |
| chr8_+_106154266 | 0.62 |
ENSMUST00000067305.8
|
Lrrc36
|
leucine rich repeat containing 36 |
| chr13_-_74642055 | 0.58 |
ENSMUST00000202645.4
ENSMUST00000221173.2 |
Zfp825
|
zinc finger protein 825 |
| chr4_-_156285247 | 0.58 |
ENSMUST00000085425.6
|
Isg15
|
ISG15 ubiquitin-like modifier |
| chr17_-_24428351 | 0.58 |
ENSMUST00000024931.6
|
Ntn3
|
netrin 3 |
| chr2_-_142743354 | 0.57 |
ENSMUST00000211861.2
|
Kif16b
|
kinesin family member 16B |
| chr14_+_53220658 | 0.53 |
ENSMUST00000200548.2
|
Trav9d-4
|
T cell receptor alpha variable 9D-4 |
| chr5_-_5713264 | 0.53 |
ENSMUST00000148193.2
|
Cfap69
|
cilia and flagella associated protein 69 |
| chr17_-_40519480 | 0.53 |
ENSMUST00000033585.7
|
Pgk2
|
phosphoglycerate kinase 2 |
| chr7_-_79115915 | 0.52 |
ENSMUST00000073889.14
|
Polg
|
polymerase (DNA directed), gamma |
| chr7_-_139785877 | 0.52 |
ENSMUST00000215815.2
|
Olfr524
|
olfactory receptor 524 |
| chr17_+_46739852 | 0.52 |
ENSMUST00000113465.10
ENSMUST00000024764.12 ENSMUST00000165993.2 |
Crip3
|
cysteine-rich protein 3 |
| chr17_+_19582211 | 0.52 |
ENSMUST00000176107.3
ENSMUST00000231989.2 |
Vmn2r99
|
vomeronasal 2, receptor 99 |
| chr6_-_129484070 | 0.51 |
ENSMUST00000183258.8
ENSMUST00000182784.4 ENSMUST00000032265.13 ENSMUST00000162815.2 |
Olr1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr3_-_58792633 | 0.50 |
ENSMUST00000055636.13
ENSMUST00000072551.7 ENSMUST00000051408.8 |
Clrn1
|
clarin 1 |
| chr7_-_103147246 | 0.50 |
ENSMUST00000216360.2
ENSMUST00000216075.2 |
Olfr609
|
olfactory receptor 609 |
| chr14_+_40794817 | 0.50 |
ENSMUST00000189865.7
|
Dydc1
|
DPY30 domain containing 1 |
| chr3_+_94172994 | 0.47 |
ENSMUST00000098878.4
|
Spopfm1
|
speckle-type BTB/POZ protein family member 1 |
| chr2_-_142743438 | 0.47 |
ENSMUST00000230763.2
ENSMUST00000043589.8 |
Kif16b
|
kinesin family member 16B |
| chr13_+_33376931 | 0.45 |
ENSMUST00000067198.3
|
Serpinb9d
|
serine (or cysteine) peptidase inhibitor, clade B, member 9d |
| chrX_-_91164377 | 0.44 |
ENSMUST00000088137.3
|
Mageb18
|
MAGE family member B18 |
| chr14_+_53913598 | 0.44 |
ENSMUST00000103662.6
|
Trav9-4
|
T cell receptor alpha variable 9-4 |
| chr12_+_85017671 | 0.44 |
ENSMUST00000021669.15
ENSMUST00000171040.2 |
Fcf1
|
FCF1 rRNA processing protein |
| chr16_-_58898368 | 0.44 |
ENSMUST00000216495.3
|
Olfr190
|
olfactory receptor 190 |
| chr7_+_44926925 | 0.42 |
ENSMUST00000210861.2
|
Slc6a21
|
solute carrier family 6 member 21 |
| chr6_+_116466052 | 0.41 |
ENSMUST00000203700.3
|
Olfr211
|
olfactory receptor 211 |
| chr13_+_33508060 | 0.40 |
ENSMUST00000075515.7
|
Serpinb9f
|
serine (or cysteine) peptidase inhibitor, clade B, member 9f |
| chr13_+_33668761 | 0.40 |
ENSMUST00000081927.4
|
Serpinb9g
|
serine (or cysteine) peptidase inhibitor, clade B, member 9g |
| chr19_-_13828056 | 0.40 |
ENSMUST00000208493.3
|
Olfr1501
|
olfactory receptor 1501 |
| chr14_+_64341735 | 0.40 |
ENSMUST00000022537.6
|
Prss52
|
protease, serine 52 |
| chr11_+_59340865 | 0.39 |
ENSMUST00000108777.10
ENSMUST00000045279.13 |
Jmjd4
|
jumonji domain containing 4 |
| chr11_-_59340739 | 0.38 |
ENSMUST00000136436.2
ENSMUST00000150297.2 ENSMUST00000010038.10 ENSMUST00000156146.8 ENSMUST00000132969.8 ENSMUST00000120940.8 |
Snap47
|
synaptosomal-associated protein, 47 |
| chr6_+_83011154 | 0.38 |
ENSMUST00000000707.9
ENSMUST00000101257.4 |
Loxl3
|
lysyl oxidase-like 3 |
| chr6_+_41204430 | 0.37 |
ENSMUST00000193064.2
ENSMUST00000103280.3 |
Trbv26
|
T cell receptor beta, variable 26 |
| chr2_+_89708781 | 0.36 |
ENSMUST00000111519.3
|
Olfr1257
|
olfactory receptor 1257 |
| chr1_+_171330978 | 0.35 |
ENSMUST00000081527.2
|
Alyref2
|
Aly/REF export factor 2 |
| chr7_+_11910120 | 0.35 |
ENSMUST00000062811.6
|
Vmn1r79
|
vomeronasal 1 receptor 79 |
| chr10_-_51507527 | 0.35 |
ENSMUST00000219286.2
ENSMUST00000020062.4 ENSMUST00000218684.2 |
Gprc6a
|
G protein-coupled receptor, family C, group 6, member A |
| chrX_+_148573187 | 0.35 |
ENSMUST00000112741.4
|
Gm15097
|
predicted gene 15097 |
| chr2_-_66240408 | 0.34 |
ENSMUST00000112366.8
|
Scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr7_-_28329924 | 0.34 |
ENSMUST00000159095.2
ENSMUST00000159418.8 ENSMUST00000159560.3 |
Acp7
|
acid phosphatase 7, tartrate resistant |
| chr8_-_25215856 | 0.33 |
ENSMUST00000033958.15
|
Adam3
|
a disintegrin and metallopeptidase domain 3 (cyritestin) |
| chr9_-_19795639 | 0.32 |
ENSMUST00000073765.6
|
Olfr862
|
olfactory receptor 862 |
| chr7_+_103583554 | 0.32 |
ENSMUST00000214711.2
|
Olfr632
|
olfactory receptor 632 |
| chr6_-_112364974 | 0.32 |
ENSMUST00000238755.2
ENSMUST00000060847.6 |
Ssu2
|
ssu-2 homolog (C. elegans) |
| chr3_+_92259448 | 0.31 |
ENSMUST00000068399.2
|
Sprr2e
|
small proline-rich protein 2E |
| chr7_+_17799849 | 0.31 |
ENSMUST00000032520.9
|
Ceacam12
|
carcinoembryonic antigen-related cell adhesion molecule 12 |
| chr18_+_35731735 | 0.31 |
ENSMUST00000236868.2
|
Paip2
|
polyadenylate-binding protein-interacting protein 2 |
| chr17_+_48070100 | 0.30 |
ENSMUST00000125177.2
|
Tfeb
|
transcription factor EB |
| chr9_-_36040075 | 0.30 |
ENSMUST00000168452.2
|
Gm5916
|
predicted gene 5916 |
| chr1_+_117855745 | 0.29 |
ENSMUST00000191265.2
|
Gm28168
|
predicted gene 28168 |
| chr11_-_51891575 | 0.28 |
ENSMUST00000109086.8
|
Ube2b
|
ubiquitin-conjugating enzyme E2B |
| chr7_+_17799889 | 0.28 |
ENSMUST00000108483.2
|
Ceacam12
|
carcinoembryonic antigen-related cell adhesion molecule 12 |
| chr15_-_74855264 | 0.27 |
ENSMUST00000023250.11
ENSMUST00000166694.2 |
Ly6i
|
lymphocyte antigen 6 complex, locus I |
| chr2_+_120331693 | 0.27 |
ENSMUST00000141181.9
|
Capn3
|
calpain 3 |
| chrX_+_122013241 | 0.26 |
ENSMUST00000113348.8
|
Cldn34c1
|
claudin 34C1 |
| chr10_-_129463803 | 0.26 |
ENSMUST00000204979.3
|
Olfr798
|
olfactory receptor 798 |
| chr2_+_52747855 | 0.25 |
ENSMUST00000155586.9
ENSMUST00000090952.11 ENSMUST00000127122.9 ENSMUST00000049483.14 ENSMUST00000050719.13 |
Fmnl2
|
formin-like 2 |
| chr11_-_49005701 | 0.23 |
ENSMUST00000060398.3
ENSMUST00000215553.2 ENSMUST00000109201.2 |
Olfr1396
|
olfactory receptor 1396 |
| chr10_-_33662700 | 0.23 |
ENSMUST00000223295.2
|
Sult3a2
|
sulfotransferase family 3A, member 2 |
| chr3_-_106729898 | 0.22 |
ENSMUST00000059486.3
|
Olfr266
|
olfactory receptor 266 |
| chr10_-_128904846 | 0.21 |
ENSMUST00000204515.3
|
Olfr766-ps1
|
olfactory receptor 766, pseudogene 1 |
| chr9_-_35701371 | 0.17 |
ENSMUST00000034619.2
|
Pate6
|
prostate and testis expressed 6 |
| chr1_+_78488499 | 0.16 |
ENSMUST00000012331.7
|
Mogat1
|
monoacylglycerol O-acyltransferase 1 |
| chr11_-_97666701 | 0.16 |
ENSMUST00000107576.2
|
1700001P01Rik
|
RIKEN cDNA 1700001P01 gene |
| chr17_+_8529932 | 0.16 |
ENSMUST00000154553.2
ENSMUST00000140890.3 |
Sft2d1
Gm49987
|
SFT2 domain containing 1 predicted gene, 49987 |
| chr14_+_33739013 | 0.15 |
ENSMUST00000226499.2
|
Gm5460
|
predicted gene 5460 |
| chr7_+_99384352 | 0.15 |
ENSMUST00000098264.2
|
Olfr520
|
olfactory receptor 520 |
| chr7_+_97049210 | 0.14 |
ENSMUST00000032882.9
ENSMUST00000149122.2 |
Ndufc2
|
NADH:ubiquinone oxidoreductase subunit C2 |
| chr7_-_100306160 | 0.13 |
ENSMUST00000107046.8
ENSMUST00000107045.9 ENSMUST00000139708.9 |
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr1_+_52047368 | 0.13 |
ENSMUST00000027277.7
|
Stat4
|
signal transducer and activator of transcription 4 |
| chr17_-_23783063 | 0.11 |
ENSMUST00000095606.5
|
Zfp213
|
zinc finger protein 213 |
| chr10_+_77891161 | 0.11 |
ENSMUST00000131825.8
ENSMUST00000139539.8 ENSMUST00000123940.2 |
Dnmt3l
|
DNA (cytosine-5-)-methyltransferase 3-like |
| chr6_-_129761233 | 0.11 |
ENSMUST00000118532.8
|
Klrh1
|
killer cell lectin-like receptor subfamily H, member 1 |
| chr2_-_89273344 | 0.10 |
ENSMUST00000216123.2
|
Olfr1240
|
olfactory receptor 1240 |
| chr6_+_133269179 | 0.08 |
ENSMUST00000048459.8
|
5530400C23Rik
|
RIKEN cDNA 5530400C23 gene |
| chr13_+_19374502 | 0.08 |
ENSMUST00000198330.2
ENSMUST00000103555.3 |
Trgv6
|
T cell receptor gamma, variable 6 |
| chr1_-_182169150 | 0.08 |
ENSMUST00000051431.10
|
Fbxo28
|
F-box protein 28 |
| chr7_-_28330322 | 0.06 |
ENSMUST00000040112.5
ENSMUST00000239470.2 |
Acp7
|
acid phosphatase 7, tartrate resistant |
| chr9_+_108269992 | 0.06 |
ENSMUST00000192995.6
|
1700102P08Rik
|
RIKEN cDNA 1700102P08 gene |
| chr5_-_115410941 | 0.04 |
ENSMUST00000040555.15
ENSMUST00000112096.9 ENSMUST00000112097.8 |
Rnf10
|
ring finger protein 10 |
| chr17_+_20565504 | 0.03 |
ENSMUST00000042090.8
|
Vmn2r107
|
vomeronasal 2, receptor 107 |
| chr17_+_18518361 | 0.02 |
ENSMUST00000231938.2
ENSMUST00000079206.8 ENSMUST00000231879.3 |
Vmn2r93
|
vomeronasal 2, receptor 93 |
| chr2_-_87543523 | 0.02 |
ENSMUST00000214209.2
|
Olfr1137
|
olfactory receptor 1137 |
| chrX_+_122013186 | 0.00 |
ENSMUST00000113343.8
|
Cldn34c1
|
claudin 34C1 |
| chr7_-_143102524 | 0.00 |
ENSMUST00000208093.2
ENSMUST00000209098.2 |
Nap1l4
|
nucleosome assembly protein 1-like 4 |
| chr7_-_107726656 | 0.00 |
ENSMUST00000214722.2
|
Olfr484
|
olfactory receptor 484 |
| chr1_+_117708863 | 0.00 |
ENSMUST00000188801.2
|
B020011L13Rik
|
RIKEN cDNA B020011L13 gene |
| chr18_-_33346819 | 0.00 |
ENSMUST00000119991.8
ENSMUST00000118990.2 |
Stard4
|
StAR-related lipid transfer (START) domain containing 4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 17.0 | GO:0097037 | heme export(GO:0097037) |
| 2.6 | 7.7 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 1.7 | 5.2 | GO:0090320 | chylomicron assembly(GO:0034378) regulation of chylomicron remnant clearance(GO:0090320) |
| 1.7 | 5.0 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.8 | 4.0 | GO:0045914 | regulation of sulfur amino acid metabolic process(GO:0031335) negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.7 | 2.8 | GO:1902081 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.4 | 27.4 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.4 | 3.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.3 | 6.3 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.3 | 1.9 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.3 | 1.9 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.3 | 0.8 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.2 | 0.9 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.2 | 4.6 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.2 | 1.1 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.2 | 3.7 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 0.7 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.1 | 11.8 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.1 | 2.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.4 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.1 | 3.7 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.1 | 1.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 1.4 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.9 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.1 | 0.3 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) negative regulation of histone phosphorylation(GO:0033128) |
| 0.1 | 0.4 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 0.6 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 0.6 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.3 | GO:1902477 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.1 | 2.4 | GO:0045821 | positive regulation of glycolytic process(GO:0045821) |
| 0.1 | 0.6 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.3 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.1 | 1.1 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 1.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.9 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 1.0 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.4 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 3.7 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.8 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.5 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 5.1 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 6.0 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 1.3 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.5 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 1.6 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.3 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.8 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.8 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) |
| 0.0 | 1.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 2.1 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 1.4 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 4.3 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 0.1 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 17.0 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.3 | 0.9 | GO:1990427 | stereocilia tip link(GO:0002140) myosin VII complex(GO:0031477) stereocilia tip-link density(GO:1990427) upper tip-link density(GO:1990435) |
| 0.2 | 0.5 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.1 | 7.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 3.7 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 32.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 1.7 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 1.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 2.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 2.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 3.2 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.1 | 6.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.1 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 1.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 1.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.6 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 3.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 7.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.1 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.7 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.6 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.7 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 1.3 | 4.0 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 1.2 | 4.6 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.9 | 11.8 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.9 | 6.0 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.8 | 5.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.7 | 3.7 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.5 | 6.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.5 | 2.4 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.4 | 3.4 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.4 | 44.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.3 | 6.7 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.3 | 1.9 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 0.9 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.2 | 3.8 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 1.6 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.5 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 1.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 0.8 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.1 | 0.6 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 0.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 1.8 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 6.9 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.1 | 0.7 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 8.1 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.9 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.8 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 1.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.5 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.2 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.5 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.4 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 5.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 3.8 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.2 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 2.8 | GO:0044325 | ion channel binding(GO:0044325) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 34.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.3 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 11.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 3.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 7.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 17.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 6.3 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 10.4 | REACTOME BIOLOGICAL OXIDATIONS | Genes involved in Biological oxidations |
| 0.1 | 2.5 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 0.9 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 1.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.9 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.9 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.6 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |