Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Sox21
Z-value: 0.94
Transcription factors associated with Sox21
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox21
|
ENSMUSG00000061517.9 | Sox21 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox21 | mm39_v1_chr14_-_118474404_118474448 | -0.21 | 8.3e-02 | Click! |
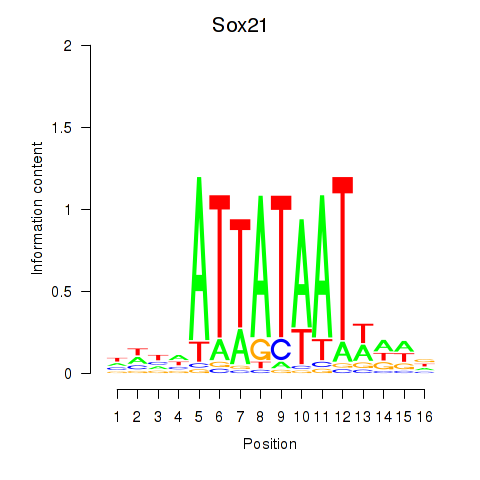
Activity profile of Sox21 motif
Sorted Z-values of Sox21 motif
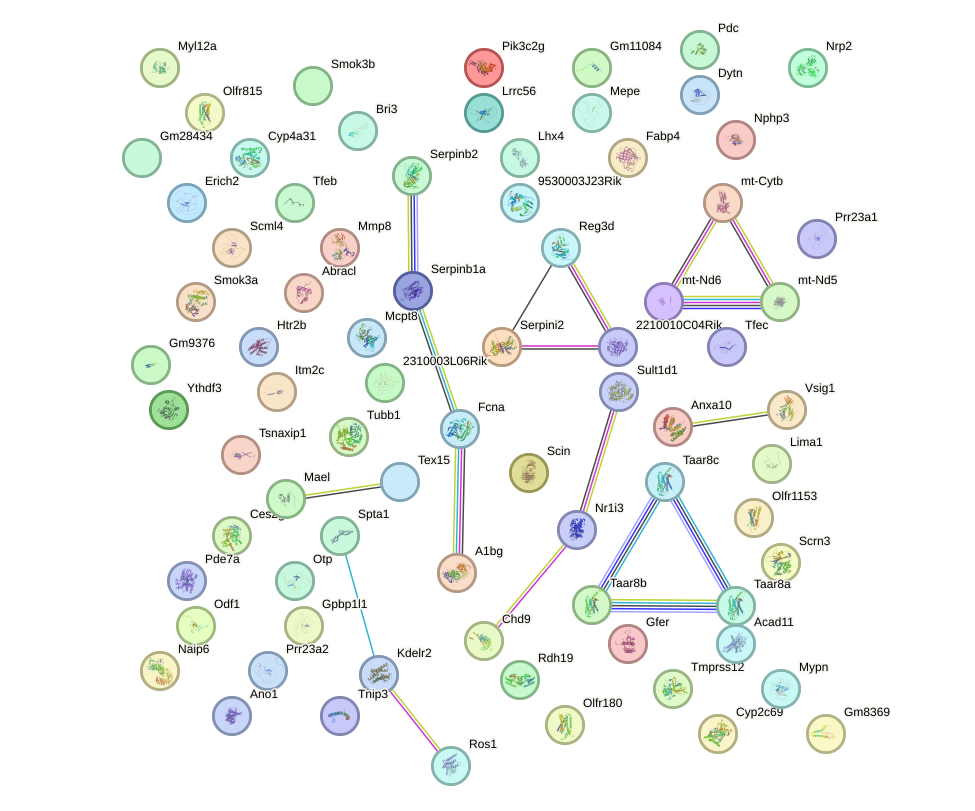
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox21
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_105688344 | 6.65 |
ENSMUST00000043183.8
|
Ces2g
|
carboxylesterase 2G |
| chr3_-_14843512 | 6.10 |
ENSMUST00000094365.11
|
Car1
|
carbonic anhydrase 1 |
| chr16_-_18904240 | 5.64 |
ENSMUST00000103746.3
|
Iglv1
|
immunoglobulin lambda variable 1 |
| chr2_+_174292471 | 4.90 |
ENSMUST00000016399.6
|
Tubb1
|
tubulin, beta 1 class VI |
| chr1_+_174000304 | 4.67 |
ENSMUST00000027817.8
|
Spta1
|
spectrin alpha, erythrocytic 1 |
| chr6_+_67586695 | 4.29 |
ENSMUST00000103303.3
|
Igkv1-135
|
immunoglobulin kappa variable 1-135 |
| chr6_+_68026941 | 4.01 |
ENSMUST00000103316.2
|
Igkv9-120
|
immunoglobulin kappa chain variable 9-120 |
| chr5_-_87716882 | 3.91 |
ENSMUST00000113314.3
|
Sult1d1
|
sulfotransferase family 1D, member 1 |
| chr6_-_16898440 | 3.67 |
ENSMUST00000031533.11
|
Tfec
|
transcription factor EC |
| chr3_-_10273628 | 3.65 |
ENSMUST00000029041.6
|
Fabp4
|
fatty acid binding protein 4, adipocyte |
| chr5_+_115061293 | 3.55 |
ENSMUST00000031540.11
ENSMUST00000112143.4 |
Oasl1
|
2'-5' oligoadenylate synthetase-like 1 |
| chr6_-_69877961 | 3.48 |
ENSMUST00000197290.2
|
Igkv5-39
|
immunoglobulin kappa variable 5-39 |
| chr10_+_23952398 | 3.30 |
ENSMUST00000051133.6
|
Taar8a
|
trace amine-associated receptor 8A |
| chr17_+_48047955 | 3.26 |
ENSMUST00000086932.10
|
Tfeb
|
transcription factor EB |
| chr14_-_56322654 | 3.22 |
ENSMUST00000015594.9
|
Mcpt8
|
mast cell protease 8 |
| chr4_-_131695135 | 3.22 |
ENSMUST00000146443.8
ENSMUST00000135579.8 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr8_-_41507808 | 3.17 |
ENSMUST00000093534.11
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr2_-_25517945 | 3.06 |
ENSMUST00000028307.9
|
Fcna
|
ficolin A |
| chr12_-_114398864 | 2.98 |
ENSMUST00000103489.2
|
Ighv6-6
|
immunoglobulin heavy variable 6-6 |
| chr8_-_62576140 | 2.85 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr19_-_34579356 | 2.78 |
ENSMUST00000168254.3
|
Ifit1bl1
|
interferon induced protein with tetratricpeptide repeats 1B like 1 |
| chr2_-_28511941 | 2.75 |
ENSMUST00000028156.8
ENSMUST00000164290.8 |
Gfi1b
|
growth factor independent 1B |
| chr9_+_7558449 | 2.67 |
ENSMUST00000018765.4
|
Mmp8
|
matrix metallopeptidase 8 |
| chrM_-_14061 | 2.59 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr3_-_30067537 | 2.42 |
ENSMUST00000108270.10
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr19_+_11469386 | 2.12 |
ENSMUST00000079855.11
|
Gm8369
|
predicted gene 8369 |
| chr2_-_72817060 | 2.12 |
ENSMUST00000112062.2
|
Gm11084
|
predicted gene 11084 |
| chrX_+_139808351 | 2.09 |
ENSMUST00000033806.5
|
Vsig1
|
V-set and immunoglobulin domain containing 1 |
| chr17_-_24915037 | 2.09 |
ENSMUST00000234235.2
|
Gfer
|
growth factor, augmenter of liver regeneration |
| chr8_+_106554353 | 2.05 |
ENSMUST00000212566.2
ENSMUST00000034365.5 |
Tsnaxip1
|
translin-associated factor X (Tsnax) interacting protein 1 |
| chr12_-_111946560 | 2.04 |
ENSMUST00000190680.2
|
Rd3l
|
retinal degeneration 3-like |
| chr13_+_95012107 | 2.03 |
ENSMUST00000022195.13
|
Otp
|
orthopedia homeobox |
| chr17_-_71305003 | 1.98 |
ENSMUST00000024846.13
ENSMUST00000232766.2 |
Myl12a
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr6_+_65567373 | 1.95 |
ENSMUST00000114236.2
|
Tnip3
|
TNFAIP3 interacting protein 3 |
| chr13_-_100454182 | 1.94 |
ENSMUST00000118574.8
|
Naip6
|
NLR family, apoptosis inhibitory protein 6 |
| chr2_+_67004178 | 1.94 |
ENSMUST00000239009.2
ENSMUST00000238912.2 |
Xirp2
|
xin actin-binding repeat containing 2 |
| chr10_-_23977810 | 1.86 |
ENSMUST00000170267.3
|
Taar8c
|
trace amine-associated receptor 8C |
| chr6_+_70549568 | 1.84 |
ENSMUST00000196940.2
ENSMUST00000103397.3 |
Igkv3-10
|
immunoglobulin kappa variable 3-10 |
| chr12_-_115083839 | 1.83 |
ENSMUST00000103521.3
|
Ighv1-50
|
immunoglobulin heavy variable 1-50 |
| chr16_+_36097313 | 1.80 |
ENSMUST00000232150.2
|
Stfa1
|
stefin A1 |
| chr3_-_75177378 | 1.78 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr6_-_78355834 | 1.77 |
ENSMUST00000089667.8
ENSMUST00000167492.4 |
Reg3d
|
regenerating islet-derived 3 delta |
| chr10_+_42736771 | 1.77 |
ENSMUST00000105494.8
|
Scml4
|
Scm polycomb group protein like 4 |
| chr3_-_16060545 | 1.75 |
ENSMUST00000194367.6
|
Gm5150
|
predicted gene 5150 |
| chr5_+_144192033 | 1.75 |
ENSMUST00000056578.7
|
Bri3
|
brain protein I3 |
| chr1_-_155617773 | 1.70 |
ENSMUST00000027740.14
|
Lhx4
|
LIM homeobox protein 4 |
| chr6_-_41012435 | 1.67 |
ENSMUST00000031931.6
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr2_+_87574098 | 1.65 |
ENSMUST00000214723.2
|
Olfr1140
|
olfactory receptor 1140 |
| chr13_-_33035150 | 1.64 |
ENSMUST00000091668.13
ENSMUST00000076352.8 |
Serpinb1a
|
serine (or cysteine) peptidase inhibitor, clade B, member 1a |
| chr1_-_97589675 | 1.60 |
ENSMUST00000053033.14
ENSMUST00000149927.2 |
Macir
|
macrophage immunometabolism regulator |
| chr6_-_130210285 | 1.53 |
ENSMUST00000032286.7
|
Klra7
|
killer cell lectin-like receptor, subfamily A, member 7 |
| chr15_-_99717956 | 1.47 |
ENSMUST00000109024.9
|
Lima1
|
LIM domain and actin binding 1 |
| chr8_+_91681898 | 1.44 |
ENSMUST00000209746.2
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr9_+_103940575 | 1.34 |
ENSMUST00000120854.8
|
Acad11
|
acyl-Coenzyme A dehydrogenase family, member 11 |
| chr1_-_86039692 | 1.33 |
ENSMUST00000027431.7
|
Htr2b
|
5-hydroxytryptamine (serotonin) receptor 2B |
| chr10_-_23968192 | 1.32 |
ENSMUST00000092654.4
|
Taar8b
|
trace amine-associated receptor 8B |
| chr5_+_76631887 | 1.32 |
ENSMUST00000191515.8
|
Gm7271
|
predicted gene 7271 |
| chrM_+_11735 | 1.25 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr8_+_34006758 | 1.21 |
ENSMUST00000149399.8
|
Tex15
|
testis expressed gene 15 |
| chr10_+_42736539 | 1.18 |
ENSMUST00000157071.8
|
Scml4
|
Scm polycomb group protein like 4 |
| chr15_+_38219447 | 1.18 |
ENSMUST00000081966.5
|
Odf1
|
outer dense fiber of sperm tails 1 |
| chr16_+_36097505 | 1.18 |
ENSMUST00000042097.11
|
Stfa1
|
stefin A1 |
| chr2_+_70339157 | 1.16 |
ENSMUST00000100041.9
|
Erich2
|
glutamate rich 2 |
| chr1_-_166066298 | 1.15 |
ENSMUST00000038782.4
ENSMUST00000194057.6 |
Mael
|
maelstrom spermatogenic transposon silencer |
| chr5_+_143389573 | 1.14 |
ENSMUST00000110731.4
|
Kdelr2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr3_-_93850850 | 1.12 |
ENSMUST00000193529.2
|
Tdpoz9-ps1
|
TD and POZ domain containing 9, pseudogene 1 |
| chr10_-_17898838 | 1.11 |
ENSMUST00000220433.2
|
Abracl
|
ABRA C-terminal like |
| chr3_-_93962192 | 1.10 |
ENSMUST00000180746.2
|
Tdpoz9
|
TD and POZ domain containing 9 |
| chr1_+_150195158 | 1.09 |
ENSMUST00000165062.8
ENSMUST00000191228.7 ENSMUST00000186572.7 ENSMUST00000185698.2 |
Pdc
|
phosducin |
| chr7_+_140774962 | 1.08 |
ENSMUST00000047093.11
|
Lrrc56
|
leucine rich repeat containing 56 |
| chr7_-_104250951 | 1.08 |
ENSMUST00000216750.2
ENSMUST00000215538.2 |
Olfr655
|
olfactory receptor 655 |
| chr10_-_17898938 | 1.08 |
ENSMUST00000220110.2
|
Abracl
|
ABRA C-terminal like |
| chr1_+_62742444 | 1.06 |
ENSMUST00000102822.9
ENSMUST00000075144.12 |
Nrp2
|
neuropilin 2 |
| chr6_+_67701864 | 1.04 |
ENSMUST00000103304.3
|
Igkv1-133
|
immunoglobulin kappa variable 1-133 |
| chr1_-_63726062 | 1.03 |
ENSMUST00000090313.5
|
Dytn
|
dystrotelin |
| chrM_+_14138 | 1.03 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chr17_-_32639936 | 1.02 |
ENSMUST00000170392.9
ENSMUST00000237165.2 ENSMUST00000235892.2 ENSMUST00000114455.3 |
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr5_+_88117307 | 0.99 |
ENSMUST00000007601.4
ENSMUST00000187738.2 |
2310003L06Rik
Gm28434
|
RIKEN cDNA 2310003L06 gene predicted gene 28434 |
| chr3_-_19319123 | 0.96 |
ENSMUST00000121951.2
|
Pde7a
|
phosphodiesterase 7A |
| chr9_+_98738547 | 0.95 |
ENSMUST00000071302.3
|
Prr23a2
|
proline rich 23A, member 2 |
| chr5_+_104473195 | 0.95 |
ENSMUST00000066207.4
|
Mepe
|
matrix extracellular phosphoglycoprotein with ASARM motif (bone) |
| chr7_-_144232586 | 0.93 |
ENSMUST00000131731.2
|
Ano1
|
anoctamin 1, calcium activated chloride channel |
| chr6_+_139564196 | 0.92 |
ENSMUST00000188066.2
ENSMUST00000190962.7 |
Pik3c2g
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
| chr10_-_129738595 | 0.91 |
ENSMUST00000071557.2
|
Olfr815
|
olfactory receptor 815 |
| chr10_-_117074501 | 0.91 |
ENSMUST00000159193.8
ENSMUST00000020392.5 |
9530003J23Rik
|
RIKEN cDNA 9530003J23 gene |
| chr6_+_42904167 | 0.90 |
ENSMUST00000101461.2
|
Olfr446
|
olfactory receptor 446 |
| chr10_-_17898977 | 0.90 |
ENSMUST00000020002.9
|
Abracl
|
ABRA C-terminal like |
| chr9_+_103940879 | 0.90 |
ENSMUST00000047799.13
ENSMUST00000189998.3 |
Acad11
|
acyl-Coenzyme A dehydrogenase family, member 11 |
| chr12_-_40184174 | 0.89 |
ENSMUST00000078481.14
ENSMUST00000002640.6 |
Scin
|
scinderin |
| chr16_-_58738861 | 0.89 |
ENSMUST00000171656.3
ENSMUST00000205883.2 |
Olfr180
|
olfactory receptor 180 |
| chr19_-_39875192 | 0.88 |
ENSMUST00000168838.3
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69 |
| chr9_+_98724556 | 0.86 |
ENSMUST00000170349.3
|
Prr23a1
|
proline rich 23A, member 1 |
| chr5_+_138019523 | 0.85 |
ENSMUST00000171498.3
ENSMUST00000085886.3 |
Smok3a
|
sperm motility kinase 3A |
| chr4_+_116414855 | 0.84 |
ENSMUST00000030460.15
|
Gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr16_-_58581603 | 0.83 |
ENSMUST00000095991.3
|
Olfr172
|
olfactory receptor 172 |
| chr2_-_87543523 | 0.83 |
ENSMUST00000214209.2
|
Olfr1137
|
olfactory receptor 1137 |
| chr3_-_37286714 | 0.83 |
ENSMUST00000161015.2
ENSMUST00000029273.8 |
Il21
|
interleukin 21 |
| chr10_-_108535970 | 0.80 |
ENSMUST00000218023.2
|
Gm5136
|
predicted gene 5136 |
| chr10_-_63039709 | 0.80 |
ENSMUST00000095580.3
|
Mypn
|
myopalladin |
| chr2_-_86941996 | 0.80 |
ENSMUST00000213978.2
|
Olfr259
|
olfactory receptor 259 |
| chr12_-_115206715 | 0.80 |
ENSMUST00000103527.2
ENSMUST00000194071.2 |
Ighv1-56
|
immunoglobulin heavy variable 1-56 |
| chr15_-_60793115 | 0.79 |
ENSMUST00000096418.5
|
A1bg
|
alpha-1-B glycoprotein |
| chr2_+_73142945 | 0.79 |
ENSMUST00000090811.11
ENSMUST00000112050.2 |
Scrn3
|
secernin 3 |
| chr10_-_52071052 | 0.77 |
ENSMUST00000218452.2
|
Ros1
|
Ros1 proto-oncogene |
| chr1_+_107439145 | 0.77 |
ENSMUST00000009356.11
ENSMUST00000064916.9 |
Serpinb2
|
serine (or cysteine) peptidase inhibitor, clade B, member 2 |
| chr2_+_85551751 | 0.76 |
ENSMUST00000055517.3
|
Olfr1009
|
olfactory receptor 1009 |
| chr7_-_85971258 | 0.74 |
ENSMUST00000044256.6
|
Olfr308
|
olfactory receptor 308 |
| chr4_+_115420876 | 0.74 |
ENSMUST00000126645.8
ENSMUST00000030480.4 |
Cyp4a31
|
cytochrome P450, family 4, subfamily a, polypeptide 31 |
| chr17_+_29487881 | 0.73 |
ENSMUST00000234845.2
ENSMUST00000235038.2 ENSMUST00000235050.2 ENSMUST00000120346.9 ENSMUST00000234377.2 ENSMUST00000235074.2 ENSMUST00000235040.2 ENSMUST00000234256.2 ENSMUST00000234459.2 |
BC004004
|
cDNA sequence BC004004 |
| chr15_+_100178718 | 0.72 |
ENSMUST00000096200.6
|
Tmprss12
|
transmembrane (C-terminal) protease, serine 12 |
| chr2_+_87726521 | 0.72 |
ENSMUST00000052300.2
|
Olfr1153
|
olfactory receptor 1153 |
| chr1_+_177635997 | 0.72 |
ENSMUST00000194528.3
|
Catspere1
|
cation channel sperm associated auxiliary subunit epsilon 1 |
| chr14_+_118504372 | 0.71 |
ENSMUST00000171107.3
|
Gm9376
|
predicted gene 9376 |
| chr10_+_127685785 | 0.71 |
ENSMUST00000077530.3
|
Rdh19
|
retinol dehydrogenase 19 |
| chr3_+_16237371 | 0.70 |
ENSMUST00000108345.9
ENSMUST00000191774.6 ENSMUST00000108346.5 |
Ythdf3
|
YTH N6-methyladenosine RNA binding protein 3 |
| chr7_-_142253247 | 0.70 |
ENSMUST00000105934.8
|
Ins2
|
insulin II |
| chr8_+_34007333 | 0.70 |
ENSMUST00000124496.8
|
Tex15
|
testis expressed gene 15 |
| chr2_+_87725306 | 0.70 |
ENSMUST00000217436.2
|
Olfr1153
|
olfactory receptor 1153 |
| chr17_+_29487762 | 0.69 |
ENSMUST00000064709.13
ENSMUST00000234711.2 |
BC004004
|
cDNA sequence BC004004 |
| chr3_-_93583617 | 0.69 |
ENSMUST00000181541.2
|
Tdpoz1
|
TD and POZ domain containing 1 |
| chr15_-_60696790 | 0.69 |
ENSMUST00000100635.5
|
Lratd2
|
LRAT domain containing 1 |
| chr5_-_113862810 | 0.69 |
ENSMUST00000047891.6
ENSMUST00000162388.8 |
1700069L16Rik
|
RIKEN cDNA 1700069L16 gene |
| chr10_+_42736644 | 0.68 |
ENSMUST00000105495.8
|
Scml4
|
Scm polycomb group protein like 4 |
| chr6_+_83985495 | 0.67 |
ENSMUST00000113821.8
|
Dysf
|
dysferlin |
| chr16_+_32066037 | 0.65 |
ENSMUST00000141820.8
ENSMUST00000178573.8 ENSMUST00000023474.4 ENSMUST00000135289.2 |
Wdr53
|
WD repeat domain 53 |
| chr11_+_49397551 | 0.64 |
ENSMUST00000213776.2
|
Olfr1384
|
olfactory receptor 1384 |
| chr8_+_43633552 | 0.64 |
ENSMUST00000209200.2
ENSMUST00000210136.2 ENSMUST00000209872.2 |
Triml2
|
tripartite motif family-like 2 |
| chr7_+_106740521 | 0.64 |
ENSMUST00000210474.2
|
Olfr716
|
olfactory receptor 716 |
| chr6_+_67816777 | 0.64 |
ENSMUST00000200578.5
ENSMUST00000103308.3 |
Igkv9-129
|
immunoglobulin kappa variable 9-129 |
| chr7_-_45367382 | 0.63 |
ENSMUST00000107737.11
|
Sphk2
|
sphingosine kinase 2 |
| chr17_-_37880331 | 0.63 |
ENSMUST00000208832.2
ENSMUST00000174798.3 |
Olfr112
|
olfactory receptor 112 |
| chr2_+_111327525 | 0.63 |
ENSMUST00000121345.4
|
Olfr1291-ps1
|
olfactory receptor 1291, pseudogene 1 |
| chr9_+_103917821 | 0.62 |
ENSMUST00000216593.2
ENSMUST00000147249.3 |
Nphp3
Gm28305
|
nephronophthisis 3 (adolescent) predicted gene 28305 |
| chr13_-_21330037 | 0.62 |
ENSMUST00000216039.3
|
Olfr1368
|
olfactory receptor 1368 |
| chr8_-_18791557 | 0.62 |
ENSMUST00000033846.7
|
Angpt2
|
angiopoietin 2 |
| chr4_-_52936281 | 0.62 |
ENSMUST00000217546.2
|
Olfr271-ps1
|
olfactory receptor 271, pseudogene 1 |
| chr2_+_88505972 | 0.61 |
ENSMUST00000216767.2
ENSMUST00000213893.2 |
Olfr1193
|
olfactory receptor 1193 |
| chr9_+_37714354 | 0.61 |
ENSMUST00000215287.2
|
Olfr876
|
olfactory receptor 876 |
| chr16_+_58967409 | 0.61 |
ENSMUST00000216957.3
|
Olfr195
|
olfactory receptor 195 |
| chr16_-_45975440 | 0.60 |
ENSMUST00000059524.7
|
Gm4737
|
predicted gene 4737 |
| chr11_+_69855584 | 0.60 |
ENSMUST00000108597.8
ENSMUST00000060651.6 ENSMUST00000108596.8 |
Cldn7
|
claudin 7 |
| chr13_+_23431304 | 0.59 |
ENSMUST00000235331.2
ENSMUST00000236495.2 ENSMUST00000238002.2 |
Vmn1r223
|
vomeronasal 1 receptor 223 |
| chr10_+_129403524 | 0.59 |
ENSMUST00000204820.3
|
Olfr794
|
olfactory receptor 794 |
| chr8_-_129534379 | 0.59 |
ENSMUST00000216104.2
|
Ccdc7a
|
coiled-coil domain containing 7A |
| chr2_-_147028309 | 0.59 |
ENSMUST00000067075.7
|
Nkx2-2
|
NK2 homeobox 2 |
| chr11_+_87590720 | 0.58 |
ENSMUST00000040089.5
|
Rnf43
|
ring finger protein 43 |
| chr7_-_45367314 | 0.58 |
ENSMUST00000210060.2
|
Sphk2
|
sphingosine kinase 2 |
| chr12_-_30181898 | 0.57 |
ENSMUST00000155263.2
|
Tpo
|
thyroid peroxidase |
| chr19_+_11495858 | 0.57 |
ENSMUST00000025580.10
|
Ms4a6b
|
membrane-spanning 4-domains, subfamily A, member 6B |
| chr13_-_99481160 | 0.56 |
ENSMUST00000022153.8
|
Ptcd2
|
pentatricopeptide repeat domain 2 |
| chr19_+_40078132 | 0.56 |
ENSMUST00000068094.13
ENSMUST00000080171.3 |
Cyp2c50
|
cytochrome P450, family 2, subfamily c, polypeptide 50 |
| chr14_+_96118660 | 0.55 |
ENSMUST00000228913.2
ENSMUST00000045892.3 |
Spertl
|
spermatid associated like |
| chr10_+_129306867 | 0.55 |
ENSMUST00000213222.2
|
Olfr788
|
olfactory receptor 788 |
| chr17_+_13449098 | 0.55 |
ENSMUST00000162940.3
|
Smok2b
|
sperm motility kinase 2B |
| chr19_+_44481901 | 0.54 |
ENSMUST00000041163.5
|
Wnt8b
|
wingless-type MMTV integration site family, member 8B |
| chr7_+_11770679 | 0.53 |
ENSMUST00000228213.2
ENSMUST00000226525.2 ENSMUST00000227320.2 |
Vmn1r77
|
vomeronasal 1 receptor 77 |
| chr11_-_73290321 | 0.53 |
ENSMUST00000131253.2
ENSMUST00000120303.9 |
Olfr1
|
olfactory receptor 1 |
| chrX_+_105626790 | 0.53 |
ENSMUST00000101296.9
ENSMUST00000101297.4 |
Gm5127
|
predicted gene 5127 |
| chr8_-_112120442 | 0.52 |
ENSMUST00000038475.9
|
Fa2h
|
fatty acid 2-hydroxylase |
| chr4_+_116415251 | 0.51 |
ENSMUST00000106475.2
|
Gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr2_-_37007795 | 0.51 |
ENSMUST00000213817.2
ENSMUST00000215927.2 |
Olfr362
|
olfactory receptor 362 |
| chr2_-_88887708 | 0.49 |
ENSMUST00000213609.3
|
Olfr1218
|
olfactory receptor 1218 |
| chr2_+_85835884 | 0.49 |
ENSMUST00000111589.3
|
Olfr1032
|
olfactory receptor 1032 |
| chr6_-_58418303 | 0.49 |
ENSMUST00000228577.2
ENSMUST00000227466.2 |
Vmn1r30
|
vomeronasal 1 receptor 30 |
| chr14_+_26414422 | 0.48 |
ENSMUST00000022433.12
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr3_-_82957104 | 0.47 |
ENSMUST00000048246.5
|
Fgb
|
fibrinogen beta chain |
| chr15_-_103338671 | 0.46 |
ENSMUST00000129837.2
ENSMUST00000146675.8 ENSMUST00000153930.8 ENSMUST00000141364.8 |
Gtsf1
|
gametocyte specific factor 1 |
| chr5_+_146769700 | 0.46 |
ENSMUST00000035983.12
|
Rpl21
|
ribosomal protein L21 |
| chr7_-_109322993 | 0.46 |
ENSMUST00000106735.9
ENSMUST00000033334.5 |
BC051019
|
cDNA sequence BC051019 |
| chr11_+_101221431 | 0.45 |
ENSMUST00000103105.10
|
Aoc3
|
amine oxidase, copper containing 3 |
| chr16_-_22848153 | 0.45 |
ENSMUST00000232459.2
|
Kng2
|
kininogen 2 |
| chr2_-_111942878 | 0.45 |
ENSMUST00000217078.2
|
Olfr1315-ps1
|
olfactory receptor 1315, pseudogene 1 |
| chr11_+_11414256 | 0.43 |
ENSMUST00000020410.11
|
Spata48
|
spermatogenesis associated 48 |
| chr16_-_58850641 | 0.43 |
ENSMUST00000216415.2
ENSMUST00000206463.3 |
Olfr186
|
olfactory receptor 186 |
| chrX_+_110808231 | 0.42 |
ENSMUST00000207546.2
|
Gm45194
|
predicted gene 45194 |
| chr18_+_4993795 | 0.42 |
ENSMUST00000153016.8
|
Svil
|
supervillin |
| chr3_+_138047536 | 0.41 |
ENSMUST00000199673.6
|
Adh6b
|
alcohol dehydrogenase 6B (class V) |
| chr2_-_111100733 | 0.40 |
ENSMUST00000099619.6
|
Olfr1277
|
olfactory receptor 1277 |
| chr7_+_106507208 | 0.40 |
ENSMUST00000215949.2
ENSMUST00000207492.3 |
Olfr1532-ps1
|
olfactory receptor 1532, pseudogene 1 |
| chr13_+_93810911 | 0.40 |
ENSMUST00000048001.8
|
Dmgdh
|
dimethylglycine dehydrogenase precursor |
| chr4_+_138503046 | 0.40 |
ENSMUST00000030528.9
|
Pla2g2d
|
phospholipase A2, group IID |
| chr2_+_88470886 | 0.40 |
ENSMUST00000217379.2
ENSMUST00000120598.3 |
Olfr1191-ps1
|
olfactory receptor 1191, pseudogene 1 |
| chr5_-_87638728 | 0.40 |
ENSMUST00000147854.6
|
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr3_+_19562753 | 0.40 |
ENSMUST00000118968.8
|
Dnajc5b
|
DnaJ heat shock protein family (Hsp40) member C5 beta |
| chr2_+_87686206 | 0.39 |
ENSMUST00000217376.2
|
Olfr1151
|
olfactory receptor 1151 |
| chr13_-_22407330 | 0.39 |
ENSMUST00000228303.2
ENSMUST00000227457.2 ENSMUST00000227562.2 |
Vmn1r193
|
vomeronasal 1 receptor 193 |
| chr9_-_38976247 | 0.39 |
ENSMUST00000215049.2
|
Olfr937
|
olfactory receptor 937 |
| chr6_+_67736650 | 0.39 |
ENSMUST00000103305.2
|
Igkv1-132
|
immunoglobulin kappa variable 1-132 |
| chr19_+_12177998 | 0.38 |
ENSMUST00000213759.2
|
Olfr1431
|
olfactory receptor 1431 |
| chr9_+_38026988 | 0.37 |
ENSMUST00000072290.5
|
Olfr889
|
olfactory receptor 889 |
| chr9_-_21996693 | 0.35 |
ENSMUST00000179422.8
ENSMUST00000098937.10 ENSMUST00000177967.2 ENSMUST00000180180.8 |
Ecsit
|
ECSIT signalling integrator |
| chr1_-_75200298 | 0.35 |
ENSMUST00000179573.2
|
A630095N17Rik
|
RIKEN cDNA A630095N17 gene |
| chr2_+_87284306 | 0.35 |
ENSMUST00000213366.3
|
Olfr1126
|
olfactory receptor 1126 |
| chrX_-_101909959 | 0.35 |
ENSMUST00000048061.13
|
1700031F05Rik
|
RIKEN cDNA 1700031F05 gene |
| chr7_-_141710850 | 0.35 |
ENSMUST00000209599.2
|
Gm29735
|
predicted gene, 29735 |
| chr3_+_94350622 | 0.35 |
ENSMUST00000029786.14
ENSMUST00000196143.2 |
Mrpl9
|
mitochondrial ribosomal protein L9 |
| chr15_+_102378966 | 0.34 |
ENSMUST00000077037.13
ENSMUST00000229102.2 ENSMUST00000229618.2 ENSMUST00000229275.2 ENSMUST00000231089.2 ENSMUST00000229802.2 ENSMUST00000229854.2 ENSMUST00000108838.5 |
Pcbp2
|
poly(rC) binding protein 2 |
| chr8_-_88686188 | 0.34 |
ENSMUST00000109655.9
|
Zfp423
|
zinc finger protein 423 |
| chr2_-_86970644 | 0.34 |
ENSMUST00000152758.4
|
Olfr1110
|
olfactory receptor 1110 |
| chr14_+_51648458 | 0.34 |
ENSMUST00000022438.12
|
Vmn2r88
|
vomeronasal 2, receptor 88 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:0009816 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.6 | 3.9 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.4 | 1.7 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.4 | 2.8 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.4 | 1.1 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.3 | 1.3 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.3 | 1.2 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.2 | 3.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.2 | 0.7 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.2 | 0.6 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.2 | 0.6 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.2 | 3.6 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.2 | 3.2 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.2 | 1.0 | GO:0032824 | negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.2 | 2.0 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.2 | 1.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.2 | 2.1 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.2 | 1.1 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.2 | 2.4 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 1.9 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.1 | 0.8 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.1 | 0.8 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 1.0 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.1 | 0.6 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 2.1 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 1.4 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 0.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 6.5 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 4.9 | GO:0033014 | tetrapyrrole biosynthetic process(GO:0033014) |
| 0.1 | 1.7 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.1 | 2.7 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 0.9 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 2.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.6 | GO:0048496 | determination of pancreatic left/right asymmetry(GO:0035469) maintenance of organ identity(GO:0048496) |
| 0.1 | 0.9 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 3.2 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 20.8 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 0.3 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 | 1.9 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 1.4 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.1 | 0.5 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.1 | 0.9 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.5 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 0.4 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 1.1 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 1.0 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 1.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 3.3 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.2 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.0 | 3.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 1.3 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.7 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 4.7 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.8 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.6 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.9 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.3 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.6 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.5 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.4 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 3.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 2.5 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 7.2 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 1.5 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 1.5 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.6 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.2 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 2.8 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.7 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.3 | 1.9 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.2 | 1.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.2 | 0.6 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 1.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 1.0 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.1 | 1.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 5.2 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.1 | 0.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.7 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 4.1 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 1.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.5 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 3.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 3.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.7 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 2.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.3 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 34.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 2.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 2.6 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 1.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.9 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.7 | 2.2 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.6 | 6.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.6 | 6.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.4 | 2.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.3 | 3.6 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.2 | 1.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.2 | 0.6 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.2 | 1.0 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.2 | 0.9 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.2 | 1.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.2 | 0.5 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 0.6 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.1 | 0.8 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 4.1 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.2 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 1.0 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 1.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 0.7 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.1 | 0.4 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.1 | 3.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.9 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 1.3 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.1 | 0.2 | GO:0070773 | protein-N-terminal glutamine amidohydrolase activity(GO:0070773) |
| 0.1 | 1.4 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 3.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.9 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 3.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.6 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 2.5 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 1.1 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 7.1 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 8.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.8 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 7.6 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 1.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.7 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 2.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 3.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.3 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 1.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 2.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 2.0 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 0.7 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 7.9 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.2 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 3.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 3.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 3.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 5.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.8 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 3.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.3 | PID ATM PATHWAY | ATM pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 3.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 1.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 2.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 4.7 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 2.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.5 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.8 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |