Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
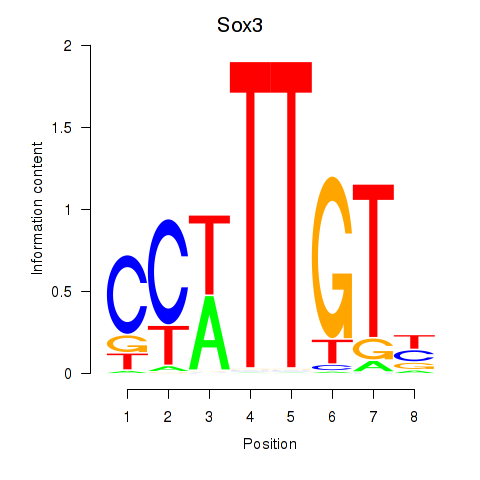
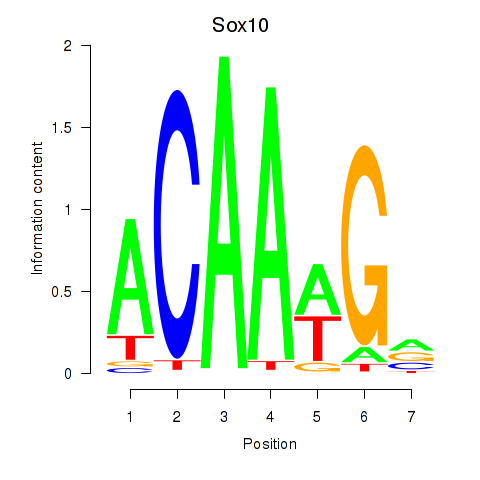
Results for Sox3_Sox10
Z-value: 2.00
Transcription factors associated with Sox3_Sox10
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox3
|
ENSMUSG00000045179.10 | Sox3 |
|
Sox10
|
ENSMUSG00000033006.11 | Sox10 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox10 | mm39_v1_chr15_-_79048674_79048696 | 0.86 | 1.3e-22 | Click! |
| Sox3 | mm39_v1_chrX_-_59937036_59937051 | 0.33 | 4.0e-03 | Click! |
Activity profile of Sox3_Sox10 motif
Sorted Z-values of Sox3_Sox10 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox3_Sox10
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.1 | 44.6 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 7.7 | 92.6 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 6.1 | 18.4 | GO:0030824 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 6.1 | 24.5 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 4.8 | 14.5 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 4.4 | 17.8 | GO:1904616 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 3.6 | 14.3 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 3.2 | 12.8 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 3.2 | 15.8 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 3.0 | 21.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 2.9 | 8.6 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 2.8 | 11.2 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 2.8 | 13.9 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 2.7 | 19.0 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 2.6 | 7.7 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 2.5 | 9.9 | GO:0098961 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 2.5 | 7.4 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 2.4 | 7.3 | GO:2000097 | negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 2.3 | 11.6 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 2.2 | 17.6 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 2.1 | 8.5 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 2.1 | 8.3 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 2.1 | 6.2 | GO:0015825 | L-serine transport(GO:0015825) |
| 2.0 | 8.0 | GO:0051866 | response to insecticide(GO:0017085) general adaptation syndrome(GO:0051866) |
| 1.8 | 9.0 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 1.8 | 10.6 | GO:0046103 | inosine biosynthetic process(GO:0046103) |
| 1.8 | 5.3 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 1.8 | 12.3 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 1.7 | 15.5 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 1.7 | 5.2 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 1.7 | 10.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 1.7 | 6.7 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 1.7 | 15.1 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.7 | 6.7 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 1.6 | 6.6 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 1.6 | 4.8 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 1.6 | 6.5 | GO:0021586 | pons maturation(GO:0021586) |
| 1.6 | 1.6 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 1.5 | 7.6 | GO:0070295 | renal water absorption(GO:0070295) |
| 1.5 | 9.0 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 1.5 | 1.5 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 1.5 | 14.6 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 1.4 | 5.5 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 1.4 | 5.5 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 1.3 | 10.6 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 1.3 | 6.5 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 1.3 | 9.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 1.3 | 3.8 | GO:0051944 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 1.3 | 5.1 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 1.2 | 20.9 | GO:1901629 | regulation of presynaptic membrane organization(GO:1901629) |
| 1.2 | 22.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 1.2 | 6.0 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 1.2 | 2.4 | GO:0052564 | response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 1.2 | 18.8 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 1.2 | 17.6 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 1.2 | 18.7 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 1.2 | 10.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 1.2 | 4.6 | GO:2000880 | dipeptide transmembrane transport(GO:0035442) regulation of oligopeptide transport(GO:0090088) regulation of dipeptide transport(GO:0090089) positive regulation of oligopeptide transport(GO:2000878) positive regulation of dipeptide transport(GO:2000880) regulation of dipeptide transmembrane transport(GO:2001148) positive regulation of dipeptide transmembrane transport(GO:2001150) |
| 1.1 | 3.4 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 1.1 | 4.4 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 1.1 | 52.5 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 1.1 | 10.6 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 1.1 | 8.4 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 1.1 | 23.1 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 1.0 | 8.3 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 1.0 | 17.4 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 1.0 | 10.2 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 1.0 | 3.0 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 1.0 | 3.0 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 1.0 | 4.0 | GO:2000657 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 1.0 | 22.0 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 1.0 | 15.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 1.0 | 9.6 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.9 | 7.6 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.9 | 2.8 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.9 | 10.4 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.9 | 11.8 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.9 | 5.4 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.9 | 3.6 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.9 | 2.7 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.9 | 8.9 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.9 | 2.6 | GO:0061744 | psychomotor behavior(GO:0036343) motor behavior(GO:0061744) |
| 0.9 | 7.9 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.9 | 4.4 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.9 | 22.8 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.9 | 13.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.9 | 4.4 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.9 | 3.4 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.8 | 4.2 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.8 | 6.4 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.8 | 4.6 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.8 | 0.8 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.8 | 3.1 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.8 | 3.1 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.8 | 6.8 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.8 | 2.3 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.8 | 4.5 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.7 | 6.7 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.7 | 4.4 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.7 | 9.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.7 | 15.3 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.7 | 2.2 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.7 | 7.7 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.7 | 4.9 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.7 | 2.8 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.7 | 4.8 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.7 | 10.3 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.7 | 8.9 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.7 | 2.7 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.7 | 12.7 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.7 | 3.3 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.7 | 7.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.7 | 18.4 | GO:1905144 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.6 | 2.6 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.6 | 2.5 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.6 | 2.5 | GO:0072023 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.6 | 2.5 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.6 | 3.1 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.6 | 13.0 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.6 | 3.1 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.6 | 22.6 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.6 | 6.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.6 | 3.6 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.6 | 1.7 | GO:1903465 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.6 | 3.5 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.6 | 1.7 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.6 | 18.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.6 | 15.5 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.6 | 8.3 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.5 | 1.1 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.5 | 9.1 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.5 | 19.8 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.5 | 2.6 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.5 | 3.6 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.5 | 3.6 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.5 | 4.6 | GO:0086018 | SA node cell action potential(GO:0086015) SA node cell to atrial cardiac muscle cell signalling(GO:0086018) SA node cell to atrial cardiac muscle cell communication(GO:0086070) |
| 0.5 | 5.1 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.5 | 1.5 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) cellular lipid biosynthetic process(GO:0097384) |
| 0.5 | 6.5 | GO:0036376 | sodium ion export from cell(GO:0036376) |
| 0.5 | 1.5 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.5 | 2.0 | GO:0015827 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.5 | 3.9 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.5 | 2.9 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.5 | 10.5 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.5 | 1.4 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.5 | 4.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.5 | 6.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.5 | 9.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.5 | 3.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.4 | 5.8 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.4 | 3.1 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 0.4 | 12.0 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.4 | 1.8 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.4 | 0.9 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.4 | 2.6 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.4 | 4.4 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.4 | 1.3 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.4 | 1.3 | GO:0021998 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) positive regulation of cardiac ventricle development(GO:1904414) |
| 0.4 | 5.5 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.4 | 4.3 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.4 | 3.4 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.4 | 1.3 | GO:0035604 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) coronal suture morphogenesis(GO:0060365) |
| 0.4 | 6.9 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.4 | 1.6 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.4 | 1.6 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.4 | 2.4 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.4 | 1.6 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.4 | 2.0 | GO:1904306 | positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.4 | 3.5 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.4 | 9.1 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.4 | 4.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.4 | 1.1 | GO:2000642 | intralumenal vesicle formation(GO:0070676) negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.4 | 1.5 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.4 | 1.9 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.4 | 1.4 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.4 | 16.7 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.4 | 14.2 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.4 | 5.3 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.3 | 2.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.3 | 1.4 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.3 | 1.0 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) positive regulation of microtubule nucleation(GO:0090063) |
| 0.3 | 0.7 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.3 | 19.0 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.3 | 4.3 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.3 | 1.6 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.3 | 1.3 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.3 | 1.0 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.3 | 1.3 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.3 | 63.8 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.3 | 2.8 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.3 | 6.4 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.3 | 4.2 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.3 | 11.7 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.3 | 11.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.3 | 0.9 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.3 | 2.3 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.3 | 1.7 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.3 | 9.3 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.3 | 0.6 | GO:0014894 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.3 | 1.4 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.3 | 1.7 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.3 | 0.8 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.3 | 0.8 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.3 | 3.9 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.3 | 3.5 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.3 | 0.8 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.3 | 1.6 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.3 | 4.6 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.3 | 0.8 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.3 | 1.3 | GO:0015965 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.3 | 31.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 4.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.2 | 6.9 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.2 | 3.7 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.2 | 1.0 | GO:0000105 | histidine biosynthetic process(GO:0000105) |
| 0.2 | 1.0 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.2 | 0.5 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.2 | 1.8 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 1.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 3.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.2 | 2.5 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.2 | 4.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 2.0 | GO:0042473 | outer ear morphogenesis(GO:0042473) |
| 0.2 | 0.2 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.2 | 5.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.2 | 0.9 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.2 | 0.9 | GO:2000293 | negative regulation of eating behavior(GO:1903999) regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.2 | 1.3 | GO:0015692 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.2 | 5.8 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.2 | 0.8 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.2 | 0.4 | GO:0045212 | neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.2 | 1.4 | GO:0061213 | positive regulation of mesonephros development(GO:0061213) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.2 | 3.5 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.2 | 5.9 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.2 | 0.8 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.2 | 6.1 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.2 | 0.6 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.2 | 1.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.2 | 2.6 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.2 | 2.7 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.2 | 47.7 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.2 | 2.5 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 10.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.2 | 1.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.2 | 3.5 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.2 | 1.3 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.2 | 5.2 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.2 | 0.5 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.2 | 6.0 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.2 | 0.8 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.2 | 1.8 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.2 | 1.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 0.8 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.2 | 1.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.2 | 5.0 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.2 | 0.8 | GO:1903751 | intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036481) regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.2 | 2.7 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 | 1.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 17.5 | GO:0035308 | negative regulation of protein dephosphorylation(GO:0035308) |
| 0.2 | 3.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.2 | 10.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.2 | 1.4 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.2 | 5.4 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 2.5 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.1 | 0.6 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.1 | 0.4 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 0.4 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 0.6 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 2.1 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 4.1 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 4.8 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 2.6 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 21.3 | GO:0007411 | axon guidance(GO:0007411) |
| 0.1 | 4.9 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 1.4 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.1 | 9.3 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 2.2 | GO:0001553 | luteinization(GO:0001553) |
| 0.1 | 2.3 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 10.4 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 1.7 | GO:0042749 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.1 | 21.7 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.1 | 2.5 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.1 | 12.0 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.1 | 0.4 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.1 | 3.9 | GO:0048512 | circadian behavior(GO:0048512) |
| 0.1 | 1.5 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 1.5 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.1 | 1.6 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.4 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.1 | 1.5 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.1 | 2.3 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 1.8 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 3.0 | GO:1903078 | positive regulation of protein localization to plasma membrane(GO:1903078) |
| 0.1 | 2.9 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 12.7 | GO:0006887 | exocytosis(GO:0006887) |
| 0.1 | 0.9 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.1 | 3.0 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 1.0 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 1.6 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 6.3 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 21.5 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 0.9 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 | 1.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 3.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 1.1 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.1 | 3.3 | GO:0008088 | axo-dendritic transport(GO:0008088) |
| 0.1 | 2.4 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 3.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 0.3 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 | 0.2 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.1 | 16.6 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.1 | 0.8 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.1 | 0.9 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 2.5 | GO:0034308 | primary alcohol metabolic process(GO:0034308) |
| 0.1 | 1.0 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.1 | 1.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.6 | GO:0014010 | Schwann cell proliferation(GO:0014010) |
| 0.1 | 0.4 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 0.7 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.1 | 1.2 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 1.5 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 1.3 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.1 | 0.2 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.1 | 1.1 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 0.8 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.1 | 0.7 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.1 | 2.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 3.7 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.1 | 0.4 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 2.6 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 1.3 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 0.5 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 3.1 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.1 | 0.9 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 4.1 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 1.1 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.1 | 1.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 0.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 1.9 | GO:0001658 | branching involved in ureteric bud morphogenesis(GO:0001658) |
| 0.1 | 8.9 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.1 | 1.4 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 0.7 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.6 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 8.6 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 1.8 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.4 | GO:0046784 | regulation of mRNA export from nucleus(GO:0010793) viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.5 | GO:0032986 | nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.8 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.7 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.0 | 0.3 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.4 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.7 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 3.1 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.0 | 0.1 | GO:1900062 | regulation of replicative cell aging(GO:1900062) |
| 0.0 | 0.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.3 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 3.5 | GO:0006914 | autophagy(GO:0006914) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.8 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.5 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 1.0 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 2.9 | GO:0007626 | locomotory behavior(GO:0007626) |
| 0.0 | 0.4 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.3 | GO:1901160 | serotonin metabolic process(GO:0042428) primary amino compound metabolic process(GO:1901160) |
| 0.0 | 0.7 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.3 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.1 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 1.3 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 1.1 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.4 | 67.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 4.9 | 44.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 4.4 | 21.8 | GO:0044393 | microspike(GO:0044393) |
| 4.3 | 17.3 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 4.0 | 15.8 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 3.6 | 14.5 | GO:0072534 | perineuronal net(GO:0072534) |
| 2.4 | 7.3 | GO:0090537 | CERF complex(GO:0090537) |
| 2.4 | 14.5 | GO:0044305 | calyx of Held(GO:0044305) |
| 2.2 | 8.8 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 2.2 | 8.6 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 2.1 | 25.0 | GO:0044327 | dendritic spine head(GO:0044327) |
| 1.6 | 12.9 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 1.6 | 6.5 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 1.4 | 12.9 | GO:0045298 | tubulin complex(GO:0045298) |
| 1.4 | 42.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 1.3 | 25.6 | GO:0031045 | dense core granule(GO:0031045) |
| 1.3 | 6.5 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 1.3 | 3.9 | GO:0043291 | RAVE complex(GO:0043291) |
| 1.2 | 3.6 | GO:0002095 | caveolar macromolecular signaling complex(GO:0002095) |
| 1.1 | 14.3 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 1.1 | 5.4 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 1.1 | 2.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 1.0 | 4.9 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 1.0 | 13.4 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.8 | 4.2 | GO:0071914 | prominosome(GO:0071914) |
| 0.8 | 2.5 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.8 | 4.1 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.8 | 21.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.8 | 26.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.7 | 3.6 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.7 | 22.9 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.7 | 10.0 | GO:0060091 | kinocilium(GO:0060091) |
| 0.7 | 6.3 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.7 | 23.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.7 | 88.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.7 | 13.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.7 | 14.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.7 | 4.6 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.6 | 16.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.6 | 9.0 | GO:0016342 | catenin complex(GO:0016342) |
| 0.6 | 2.5 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.6 | 10.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.6 | 8.5 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.6 | 6.0 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.6 | 3.6 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.6 | 3.5 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.6 | 55.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.6 | 3.5 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.6 | 6.7 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.6 | 4.4 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.5 | 7.7 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.5 | 8.7 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.5 | 6.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.5 | 1.4 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.4 | 24.7 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.4 | 2.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.4 | 36.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.4 | 1.6 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.4 | 13.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.4 | 6.8 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.4 | 5.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.4 | 5.5 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.4 | 7.0 | GO:0042599 | lamellar body(GO:0042599) |
| 0.4 | 2.6 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.4 | 5.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.3 | 37.2 | GO:0034705 | potassium channel complex(GO:0034705) |
| 0.3 | 55.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.3 | 3.0 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.3 | 23.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.3 | 12.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.3 | 28.3 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.3 | 1.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.3 | 4.0 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.3 | 3.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.3 | 1.8 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.3 | 9.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.3 | 14.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.3 | 8.0 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.3 | 4.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.3 | 8.9 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.3 | 2.5 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.3 | 9.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.2 | 6.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 1.5 | GO:0071547 | piP-body(GO:0071547) |
| 0.2 | 44.8 | GO:0030426 | growth cone(GO:0030426) |
| 0.2 | 2.0 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 0.7 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.2 | 2.4 | GO:0042581 | specific granule(GO:0042581) |
| 0.2 | 1.3 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 1.3 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.2 | 17.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.2 | 26.1 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.2 | 14.0 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 0.8 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.2 | 109.5 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.2 | 25.8 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.2 | 2.2 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.2 | 0.9 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.2 | 30.6 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.2 | 34.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.2 | 3.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 2.2 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.1 | 24.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 4.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 6.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.8 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 1.8 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 1.4 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 1.4 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 0.4 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 3.5 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 0.9 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 0.9 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 1.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 0.2 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.1 | 3.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 12.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 3.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 2.1 | GO:0034702 | ion channel complex(GO:0034702) |
| 0.1 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 12.7 | GO:0045202 | synapse(GO:0045202) |
| 0.1 | 11.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.2 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 60.8 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.9 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 1.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.4 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 10.5 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.7 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.5 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 2.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 1.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.5 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.8 | 147.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 5.1 | 45.7 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 4.3 | 42.8 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 3.5 | 13.9 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 3.2 | 9.6 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 3.0 | 8.9 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 2.9 | 14.6 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 2.9 | 8.6 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 2.2 | 8.9 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 2.2 | 12.9 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 2.1 | 8.2 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 2.0 | 11.9 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 1.8 | 5.5 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 1.8 | 7.3 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 1.8 | 9.0 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 1.7 | 12.0 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 1.7 | 3.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 1.6 | 4.7 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 1.5 | 9.0 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 1.4 | 14.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 1.4 | 23.5 | GO:0005522 | profilin binding(GO:0005522) |
| 1.4 | 13.8 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 1.3 | 4.0 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 1.3 | 5.3 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 1.3 | 17.6 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 1.2 | 4.7 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 1.1 | 16.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 1.1 | 3.4 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 1.1 | 1.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 1.0 | 10.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 1.0 | 6.8 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 1.0 | 4.9 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.0 | 6.7 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.9 | 7.5 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.9 | 2.8 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.9 | 10.9 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.9 | 7.2 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.9 | 3.6 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.9 | 9.6 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.8 | 8.4 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.8 | 32.0 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.8 | 9.0 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.8 | 14.5 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.8 | 2.4 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.8 | 5.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.8 | 3.8 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.8 | 53.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.7 | 6.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.7 | 5.2 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.7 | 4.4 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.7 | 5.8 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.7 | 7.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.7 | 8.5 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.7 | 3.5 | GO:0102344 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.7 | 3.5 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.7 | 18.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.7 | 7.3 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.7 | 7.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.7 | 2.6 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.7 | 17.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.6 | 5.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.6 | 5.2 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.6 | 14.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.6 | 2.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.6 | 10.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.6 | 6.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.6 | 3.9 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.6 | 7.3 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.5 | 8.6 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.5 | 9.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.5 | 49.9 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.5 | 4.0 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.5 | 8.8 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.5 | 1.4 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.5 | 19.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.5 | 4.6 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.5 | 26.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.4 | 11.1 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.4 | 16.0 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.4 | 2.6 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.4 | 5.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.4 | 23.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.4 | 16.1 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.4 | 14.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.4 | 6.7 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.4 | 6.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.4 | 2.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.4 | 6.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.4 | 5.3 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.4 | 1.1 | GO:0016213 | linoleoyl-CoA desaturase activity(GO:0016213) |
| 0.4 | 1.5 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.4 | 12.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.4 | 4.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.4 | 41.3 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.3 | 3.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.3 | 1.7 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.3 | 1.7 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.3 | 50.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.3 | 11.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.3 | 6.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.3 | 2.0 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.3 | 2.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.3 | 1.6 | GO:0005113 | patched binding(GO:0005113) |
| 0.3 | 9.5 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.3 | 1.6 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.3 | 2.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.3 | 9.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 6.7 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.3 | 101.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.3 | 15.9 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.3 | 8.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.3 | 4.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.3 | 1.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.3 | 9.1 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.3 | 6.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.3 | 1.6 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.3 | 61.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.3 | 4.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 8.7 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.2 | 6.8 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 3.1 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.2 | 0.7 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.2 | 1.4 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.2 | 7.0 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.2 | 0.5 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 1.6 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.2 | 2.6 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 0.2 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.2 | 1.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.2 | 8.1 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.2 | 11.0 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 13.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.2 | 3.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 0.8 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.2 | 3.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 1.6 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.2 | 1.2 | GO:0016807 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.2 | 7.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.2 | 6.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.2 | 12.4 | GO:0017022 | myosin binding(GO:0017022) |
| 0.2 | 0.7 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.2 | 0.5 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.2 | 1.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.2 | 0.9 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 6.6 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.2 | 4.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.2 | 4.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 4.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 32.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.2 | 1.5 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.2 | 6.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.2 | 4.0 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.2 | 5.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 1.0 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.2 | 50.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.2 | 10.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.2 | 0.8 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.2 | 0.8 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 4.8 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 0.6 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 0.4 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.1 | 8.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 6.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 7.3 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 0.6 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 5.4 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 1.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 3.0 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 1.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 3.1 | GO:0042171 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.1 | 1.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 2.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 1.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 0.8 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 8.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.5 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 1.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 1.3 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 2.4 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.6 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 2.9 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 0.8 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 27.3 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.1 | 7.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 4.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 1.9 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 2.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 1.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 3.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 1.5 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.9 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 1.9 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.6 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.9 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 4.4 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 0.8 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 15.8 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 0.8 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 2.4 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 2.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 5.8 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 0.9 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 9.1 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 1.4 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 9.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 1.2 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 1.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 11.8 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 0.5 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 2.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.2 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.1 | 0.9 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 2.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 23.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 2.4 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 4.0 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 15.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 3.3 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 1.7 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 1.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.4 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.9 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.3 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 4.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 3.4 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.3 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 1.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.9 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 19.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.8 | 18.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.8 | 25.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.7 | 17.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.7 | 15.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.6 | 50.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.6 | 9.1 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.5 | 23.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.5 | 44.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.5 | 29.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.4 | 22.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.3 | 16.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.3 | 20.2 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.3 | 29.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.3 | 12.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.3 | 11.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 6.5 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.2 | 9.6 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.2 | 11.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 9.1 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.2 | 11.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.2 | 4.7 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.2 | 1.4 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.2 | 5.9 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.2 | 2.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 1.1 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 2.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 3.9 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 1.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 2.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 0.8 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 2.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 1.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 0.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 6.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 0.5 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 3.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 1.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 0.6 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.7 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 1.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.6 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.6 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.5 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.7 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.3 | PID CDC42 PATHWAY | CDC42 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 57.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 1.4 | 35.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 1.2 | 23.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 1.1 | 17.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.8 | 11.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.7 | 14.7 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.6 | 36.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.5 | 7.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.5 | 8.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.5 | 17.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.5 | 16.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.5 | 7.6 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.4 | 10.7 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.4 | 42.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.4 | 11.6 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.4 | 19.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.4 | 1.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.4 | 2.3 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.4 | 6.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.3 | 9.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.3 | 7.8 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.3 | 4.9 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.3 | 12.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.3 | 7.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.3 | 40.4 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.3 | 3.4 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.3 | 10.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.3 | 13.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.2 | 2.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 2.5 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.2 | 4.0 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 8.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 6.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 2.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 6.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 7.4 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.2 | 7.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 22.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 1.7 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 5.6 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 6.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 5.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 1.6 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 15.0 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.1 | 6.6 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.1 | 2.6 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.1 | 3.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 3.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 2.8 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 7.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 2.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 14.1 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 4.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.0 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 4.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 0.1 | REACTOME GAB1 SIGNALOSOME | Genes involved in GAB1 signalosome |
| 0.1 | 1.0 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.4 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.8 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 2.2 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 0.8 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.0 | 0.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.6 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.1 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |