Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
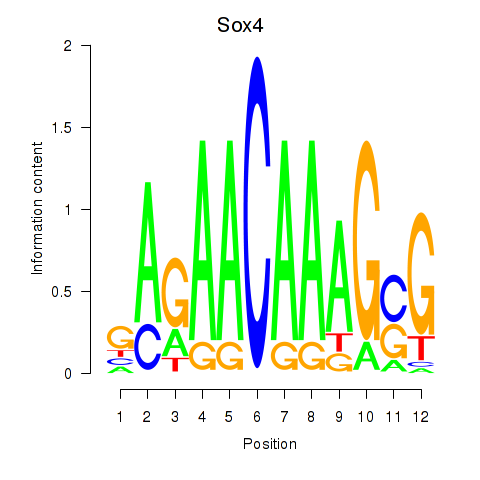
Results for Sox4
Z-value: 0.78
Transcription factors associated with Sox4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox4
|
ENSMUSG00000076431.5 | Sox4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox4 | mm39_v1_chr13_-_29137673_29137696 | 0.52 | 3.4e-06 | Click! |
Activity profile of Sox4 motif
Sorted Z-values of Sox4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_119978773 | 10.08 |
ENSMUST00000068698.15
ENSMUST00000215512.2 ENSMUST00000111627.3 ENSMUST00000093773.8 |
Mobp
|
myelin-associated oligodendrocytic basic protein |
| chr12_-_72283465 | 7.24 |
ENSMUST00000021497.16
ENSMUST00000137990.2 |
Rtn1
|
reticulon 1 |
| chr19_-_46315543 | 7.13 |
ENSMUST00000223917.2
ENSMUST00000224447.2 ENSMUST00000041391.5 ENSMUST00000096029.12 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr7_-_30614249 | 6.49 |
ENSMUST00000190950.7
ENSMUST00000187137.7 ENSMUST00000190638.7 |
Mag
|
myelin-associated glycoprotein |
| chr12_-_111638722 | 6.27 |
ENSMUST00000001304.9
|
Ckb
|
creatine kinase, brain |
| chr1_-_93029547 | 6.19 |
ENSMUST00000112958.9
ENSMUST00000186861.2 ENSMUST00000171556.8 |
Kif1a
|
kinesin family member 1A |
| chr1_-_93029532 | 6.08 |
ENSMUST00000171796.8
|
Kif1a
|
kinesin family member 1A |
| chr12_-_72455708 | 5.99 |
ENSMUST00000078505.14
|
Rtn1
|
reticulon 1 |
| chr18_+_82572595 | 5.94 |
ENSMUST00000152071.9
ENSMUST00000142850.9 ENSMUST00000080658.12 ENSMUST00000133193.9 ENSMUST00000123251.9 ENSMUST00000153478.9 ENSMUST00000075372.13 ENSMUST00000102812.12 ENSMUST00000062446.15 ENSMUST00000114674.11 ENSMUST00000132369.3 |
Mbp
|
myelin basic protein |
| chr3_-_50398027 | 5.18 |
ENSMUST00000029297.6
ENSMUST00000194462.6 |
Slc7a11
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
| chr10_-_116309764 | 5.07 |
ENSMUST00000068233.11
|
Kcnmb4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chr8_-_4309257 | 5.03 |
ENSMUST00000053252.9
|
Ctxn1
|
cortexin 1 |
| chr1_-_93029576 | 4.82 |
ENSMUST00000190723.7
|
Kif1a
|
kinesin family member 1A |
| chr5_-_146521629 | 4.40 |
ENSMUST00000200112.2
ENSMUST00000197431.2 ENSMUST00000197825.2 |
Gpr12
|
G-protein coupled receptor 12 |
| chr7_+_87233554 | 4.38 |
ENSMUST00000125009.9
|
Grm5
|
glutamate receptor, metabotropic 5 |
| chr3_+_8574420 | 4.15 |
ENSMUST00000029002.9
|
Stmn2
|
stathmin-like 2 |
| chr18_-_43192483 | 4.12 |
ENSMUST00000025377.14
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr12_+_24881582 | 4.08 |
ENSMUST00000221952.2
ENSMUST00000078902.8 ENSMUST00000110942.11 |
Mboat2
|
membrane bound O-acyltransferase domain containing 2 |
| chr12_-_81827893 | 4.04 |
ENSMUST00000035987.9
|
Map3k9
|
mitogen-activated protein kinase kinase kinase 9 |
| chr9_+_59614877 | 4.03 |
ENSMUST00000128944.8
ENSMUST00000098661.10 |
Gramd2
|
GRAM domain containing 2 |
| chr1_+_158189831 | 3.70 |
ENSMUST00000193042.6
ENSMUST00000046110.16 |
Astn1
|
astrotactin 1 |
| chr11_+_54205722 | 3.66 |
ENSMUST00000072178.11
ENSMUST00000101211.9 ENSMUST00000101213.9 ENSMUST00000064690.10 ENSMUST00000108899.8 |
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr14_+_55173696 | 3.63 |
ENSMUST00000037814.8
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr9_-_103357564 | 3.62 |
ENSMUST00000124310.5
|
Bfsp2
|
beaded filament structural protein 2, phakinin |
| chr1_-_79838897 | 3.31 |
ENSMUST00000190724.2
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chr4_+_129407374 | 3.31 |
ENSMUST00000062356.7
|
Marcksl1
|
MARCKS-like 1 |
| chr4_-_149211145 | 3.25 |
ENSMUST00000030815.3
|
Cort
|
cortistatin |
| chr17_+_12338161 | 3.20 |
ENSMUST00000024594.9
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chr15_+_30457772 | 2.83 |
ENSMUST00000228282.2
|
Ctnnd2
|
catenin (cadherin associated protein), delta 2 |
| chr14_+_14159978 | 2.81 |
ENSMUST00000137133.2
ENSMUST00000036070.15 ENSMUST00000121887.8 |
Fam107a
|
family with sequence similarity 107, member A |
| chr1_+_153541020 | 2.78 |
ENSMUST00000152114.8
ENSMUST00000111812.8 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr13_+_117356808 | 2.66 |
ENSMUST00000022242.9
|
Emb
|
embigin |
| chr18_+_37617848 | 2.63 |
ENSMUST00000053856.6
|
Pcdhb17
|
protocadherin beta 17 |
| chr14_+_55173936 | 2.59 |
ENSMUST00000227441.2
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr16_+_16964801 | 2.57 |
ENSMUST00000232479.2
ENSMUST00000232344.2 ENSMUST00000069064.7 |
Ydjc
|
YdjC homolog (bacterial) |
| chrX_+_158623460 | 2.54 |
ENSMUST00000112451.8
ENSMUST00000112453.9 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr3_-_143910926 | 2.41 |
ENSMUST00000120539.8
ENSMUST00000196264.5 |
Lmo4
|
LIM domain only 4 |
| chr6_+_71684846 | 2.37 |
ENSMUST00000212792.2
|
Reep1
|
receptor accessory protein 1 |
| chr1_-_131127825 | 2.35 |
ENSMUST00000068564.15
|
Rassf5
|
Ras association (RalGDS/AF-6) domain family member 5 |
| chr12_+_29578354 | 2.25 |
ENSMUST00000218583.2
ENSMUST00000049784.17 |
Myt1l
|
myelin transcription factor 1-like |
| chr6_+_71684872 | 2.17 |
ENSMUST00000212631.2
|
Reep1
|
receptor accessory protein 1 |
| chr12_+_24880878 | 2.15 |
ENSMUST00000222198.2
|
Mboat2
|
membrane bound O-acyltransferase domain containing 2 |
| chr2_-_165740864 | 2.07 |
ENSMUST00000136842.2
|
Zmynd8
|
zinc finger, MYND-type containing 8 |
| chr10_+_29019645 | 2.06 |
ENSMUST00000092629.4
|
Soga3
|
SOGA family member 3 |
| chr17_+_69144053 | 2.05 |
ENSMUST00000178545.3
|
Tmem200c
|
transmembrane protein 200C |
| chr14_-_61283911 | 2.05 |
ENSMUST00000111234.10
ENSMUST00000224371.2 |
Tnfrsf19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr11_+_45946800 | 1.83 |
ENSMUST00000011400.8
|
Adam19
|
a disintegrin and metallopeptidase domain 19 (meltrin beta) |
| chr4_-_91288221 | 1.82 |
ENSMUST00000102799.10
|
Elavl2
|
ELAV like RNA binding protein 1 |
| chr19_-_18978463 | 1.76 |
ENSMUST00000040153.15
ENSMUST00000112828.8 |
Rorb
|
RAR-related orphan receptor beta |
| chr18_+_37630044 | 1.67 |
ENSMUST00000059571.7
|
Pcdhb19
|
protocadherin beta 19 |
| chr6_+_137229427 | 1.65 |
ENSMUST00000077115.13
|
Ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr3_-_143908111 | 1.64 |
ENSMUST00000121796.8
|
Lmo4
|
LIM domain only 4 |
| chr3_-_143908060 | 1.57 |
ENSMUST00000121112.6
|
Lmo4
|
LIM domain only 4 |
| chr5_+_20112704 | 1.55 |
ENSMUST00000115267.7
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr1_-_131441962 | 1.52 |
ENSMUST00000185445.3
|
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr11_-_106050927 | 1.52 |
ENSMUST00000045923.10
|
Limd2
|
LIM domain containing 2 |
| chr11_-_113956996 | 1.49 |
ENSMUST00000041627.14
|
Sdk2
|
sidekick cell adhesion molecule 2 |
| chr6_-_23839136 | 1.37 |
ENSMUST00000166458.9
ENSMUST00000142913.9 ENSMUST00000069074.14 ENSMUST00000115361.9 ENSMUST00000018122.14 ENSMUST00000115356.3 |
Cadps2
|
Ca2+-dependent activator protein for secretion 2 |
| chr1_+_158189900 | 1.35 |
ENSMUST00000170718.7
|
Astn1
|
astrotactin 1 |
| chr5_+_122534900 | 1.29 |
ENSMUST00000196969.5
|
Arpc3
|
actin related protein 2/3 complex, subunit 3 |
| chr18_+_65022035 | 1.27 |
ENSMUST00000224385.3
ENSMUST00000163516.9 |
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr3_-_127631068 | 1.23 |
ENSMUST00000051737.8
ENSMUST00000200409.5 |
Ap1ar
|
adaptor-related protein complex 1 associated regulatory protein |
| chr11_-_78388560 | 1.21 |
ENSMUST00000061174.7
|
Sarm1
|
sterile alpha and HEAT/Armadillo motif containing 1 |
| chr17_-_36015484 | 1.20 |
ENSMUST00000117301.8
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr19_+_44919585 | 1.18 |
ENSMUST00000096053.5
|
Slf2
|
SMC5-SMC6 complex localization factor 2 |
| chr16_-_18885809 | 1.17 |
ENSMUST00000200211.2
|
Iglj3
|
immunoglobulin lambda joining 3 |
| chr3_-_105940130 | 1.11 |
ENSMUST00000200146.2
|
Chil5
|
chitinase-like 5 |
| chr18_+_37622518 | 1.11 |
ENSMUST00000055949.4
|
Pcdhb18
|
protocadherin beta 18 |
| chr18_+_37433852 | 1.10 |
ENSMUST00000051754.2
|
Pcdhb3
|
protocadherin beta 3 |
| chr2_-_26854283 | 1.10 |
ENSMUST00000136710.2
ENSMUST00000064244.11 ENSMUST00000114020.10 |
Rexo4
|
REX4, 3'-5' exonuclease |
| chr7_-_100164007 | 1.10 |
ENSMUST00000207405.2
|
Dnajb13
|
DnaJ heat shock protein family (Hsp40) member B13 |
| chr1_-_36982747 | 1.07 |
ENSMUST00000185964.3
|
Tmem131
|
transmembrane protein 131 |
| chr8_+_89423645 | 1.04 |
ENSMUST00000043526.15
ENSMUST00000211554.2 ENSMUST00000209532.2 ENSMUST00000209559.2 |
Cyld
|
CYLD lysine 63 deubiquitinase |
| chr11_-_78388284 | 1.04 |
ENSMUST00000108287.10
|
Sarm1
|
sterile alpha and HEAT/Armadillo motif containing 1 |
| chr5_+_20112500 | 1.00 |
ENSMUST00000101558.10
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr6_+_41139948 | 0.95 |
ENSMUST00000103275.4
|
Trbv17
|
T cell receptor beta, variable 17 |
| chr4_+_83335947 | 0.90 |
ENSMUST00000030206.10
ENSMUST00000071544.11 |
Snapc3
|
small nuclear RNA activating complex, polypeptide 3 |
| chr5_+_25452342 | 0.88 |
ENSMUST00000114950.2
|
Galnt11
|
polypeptide N-acetylgalactosaminyltransferase 11 |
| chr11_+_22462088 | 0.88 |
ENSMUST00000059319.8
|
Tmem17
|
transmembrane protein 17 |
| chr14_-_52495160 | 0.86 |
ENSMUST00000200169.6
|
Chd8
|
chromodomain helicase DNA binding protein 8 |
| chr19_+_7394951 | 0.86 |
ENSMUST00000159348.3
|
2700081O15Rik
|
RIKEN cDNA 2700081O15 gene |
| chr4_+_136013372 | 0.83 |
ENSMUST00000069195.5
ENSMUST00000130658.2 |
Zfp46
|
zinc finger protein 46 |
| chr2_+_32460868 | 0.82 |
ENSMUST00000140592.8
ENSMUST00000028151.7 |
Dpm2
|
dolichol-phosphate (beta-D) mannosyltransferase 2 |
| chr8_+_85696695 | 0.82 |
ENSMUST00000164807.2
|
Prdx2
|
peroxiredoxin 2 |
| chr16_-_90841360 | 0.81 |
ENSMUST00000118522.8
|
Paxbp1
|
PAX3 and PAX7 binding protein 1 |
| chr11_+_69737491 | 0.80 |
ENSMUST00000019605.4
|
Plscr3
|
phospholipid scramblase 3 |
| chr7_-_143102792 | 0.78 |
ENSMUST00000072727.7
ENSMUST00000207948.2 ENSMUST00000208190.2 |
Nap1l4
|
nucleosome assembly protein 1-like 4 |
| chr11_+_69737437 | 0.75 |
ENSMUST00000152566.8
ENSMUST00000108633.9 |
Plscr3
|
phospholipid scramblase 3 |
| chr3_+_89072096 | 0.74 |
ENSMUST00000121212.9
ENSMUST00000152205.5 ENSMUST00000090927.12 ENSMUST00000148265.8 ENSMUST00000121931.8 |
Clk2
|
CDC-like kinase 2 |
| chr17_-_36008863 | 0.74 |
ENSMUST00000146472.8
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr15_+_76131020 | 0.70 |
ENSMUST00000229380.2
|
Grina
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr5_+_36050663 | 0.69 |
ENSMUST00000064571.11
|
Afap1
|
actin filament associated protein 1 |
| chr5_-_136227760 | 0.66 |
ENSMUST00000149151.2
ENSMUST00000151786.8 |
Prkrip1
|
Prkr interacting protein 1 (IL11 inducible) |
| chr1_-_55066629 | 0.62 |
ENSMUST00000027127.14
|
Sf3b1
|
splicing factor 3b, subunit 1 |
| chr18_-_36899245 | 0.62 |
ENSMUST00000061522.8
|
Dnd1
|
DND microRNA-mediated repression inhibitor 1 |
| chr3_-_95041246 | 0.61 |
ENSMUST00000172572.9
ENSMUST00000173462.3 |
Scnm1
|
sodium channel modifier 1 |
| chr1_-_64161415 | 0.60 |
ENSMUST00000135075.2
|
Klf7
|
Kruppel-like factor 7 (ubiquitous) |
| chr11_+_69737200 | 0.60 |
ENSMUST00000108632.8
|
Plscr3
|
phospholipid scramblase 3 |
| chr13_-_59930059 | 0.57 |
ENSMUST00000225581.2
|
Gm49354
|
predicted gene, 49354 |
| chr19_+_34899778 | 0.51 |
ENSMUST00000223907.2
|
Kif20b
|
kinesin family member 20B |
| chrX_-_101114906 | 0.51 |
ENSMUST00000188731.2
|
Rtl5
|
retrotransposon Gag like 5 |
| chr8_+_85696396 | 0.50 |
ENSMUST00000109733.8
|
Prdx2
|
peroxiredoxin 2 |
| chr1_-_153363354 | 0.49 |
ENSMUST00000186380.7
ENSMUST00000188345.2 ENSMUST00000042141.12 |
Dhx9
|
DEAH (Asp-Glu-Ala-His) box polypeptide 9 |
| chr17_-_36015496 | 0.47 |
ENSMUST00000134995.8
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr19_+_34899752 | 0.44 |
ENSMUST00000087341.7
|
Kif20b
|
kinesin family member 20B |
| chr7_+_66393252 | 0.43 |
ENSMUST00000066475.11
|
Cers3
|
ceramide synthase 3 |
| chr17_+_37504783 | 0.43 |
ENSMUST00000038844.7
|
Ubd
|
ubiquitin D |
| chr19_-_4384029 | 0.41 |
ENSMUST00000176653.2
|
Kdm2a
|
lysine (K)-specific demethylase 2A |
| chr15_-_84740243 | 0.39 |
ENSMUST00000159939.8
|
Phf21b
|
PHD finger protein 21B |
| chr8_+_85696453 | 0.36 |
ENSMUST00000125893.8
|
Prdx2
|
peroxiredoxin 2 |
| chr16_-_31094095 | 0.35 |
ENSMUST00000060188.14
|
Ppp1r2
|
protein phosphatase 1, regulatory inhibitor subunit 2 |
| chr8_+_95721378 | 0.32 |
ENSMUST00000212956.2
ENSMUST00000212531.2 |
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr17_+_28794615 | 0.31 |
ENSMUST00000232862.2
ENSMUST00000080780.8 |
Lhfpl5
|
lipoma HMGIC fusion partner-like 5 |
| chr12_-_75678092 | 0.31 |
ENSMUST00000238938.2
|
Rplp2-ps1
|
ribosomal protein, large P2, pseudogene 1 |
| chr5_+_110987839 | 0.31 |
ENSMUST00000200172.2
ENSMUST00000066160.3 |
Chek2
|
checkpoint kinase 2 |
| chr12_-_110662723 | 0.29 |
ENSMUST00000021698.13
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr11_+_101333238 | 0.27 |
ENSMUST00000107249.8
|
Rpl27
|
ribosomal protein L27 |
| chr15_+_76130947 | 0.27 |
ENSMUST00000229772.2
ENSMUST00000230347.2 ENSMUST00000023225.8 |
Grina
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr17_-_79662514 | 0.27 |
ENSMUST00000068958.9
|
Cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr19_+_5790918 | 0.26 |
ENSMUST00000081496.6
|
Ltbp3
|
latent transforming growth factor beta binding protein 3 |
| chr14_+_102077937 | 0.22 |
ENSMUST00000159026.8
|
Lmo7
|
LIM domain only 7 |
| chr8_+_95720864 | 0.21 |
ENSMUST00000212141.2
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr11_+_101333115 | 0.21 |
ENSMUST00000077856.13
|
Rpl27
|
ribosomal protein L27 |
| chr7_+_126832399 | 0.19 |
ENSMUST00000056232.7
|
Zfp553
|
zinc finger protein 553 |
| chr7_+_99115211 | 0.18 |
ENSMUST00000037359.9
|
Klhl35
|
kelch-like 35 |
| chr3_-_75389047 | 0.17 |
ENSMUST00000193989.4
ENSMUST00000203169.3 |
Wdr49
|
WD repeat domain 49 |
| chr12_-_110662677 | 0.15 |
ENSMUST00000124156.8
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr15_-_5273645 | 0.15 |
ENSMUST00000120563.2
|
Ptger4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr8_+_95721019 | 0.14 |
ENSMUST00000212976.2
ENSMUST00000212995.2 |
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr19_-_32080496 | 0.13 |
ENSMUST00000235213.2
ENSMUST00000236504.2 |
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr16_-_13977084 | 0.12 |
ENSMUST00000090300.6
|
Marf1
|
meiosis regulator and mRNA stability 1 |
| chr8_-_85696040 | 0.12 |
ENSMUST00000214133.2
ENSMUST00000147812.8 |
Gm49661
Rnaseh2a
|
predicted gene, 49661 ribonuclease H2, large subunit |
| chr7_+_48895879 | 0.10 |
ENSMUST00000064395.13
|
Nav2
|
neuron navigator 2 |
| chr6_-_93769426 | 0.08 |
ENSMUST00000204788.2
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr16_+_37909363 | 0.01 |
ENSMUST00000023507.13
|
Gsk3b
|
glycogen synthase kinase 3 beta |
| chr7_-_103502404 | 0.00 |
ENSMUST00000033229.5
|
Hbb-y
|
hemoglobin Y, beta-like embryonic chain |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 5.9 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 1.5 | 4.4 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 1.1 | 3.3 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 1.0 | 4.2 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.8 | 10.1 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.7 | 3.7 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.7 | 17.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.6 | 5.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.5 | 5.2 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.4 | 2.5 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.4 | 1.6 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.3 | 6.3 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.3 | 2.1 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.3 | 6.5 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.3 | 5.0 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.3 | 1.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.3 | 0.3 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.2 | 4.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.2 | 5.6 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.2 | 2.7 | GO:0035879 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.2 | 4.5 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.2 | 1.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.2 | 2.8 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.2 | 1.2 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) protein localization to site of double-strand break(GO:1990166) |
| 0.2 | 7.1 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 2.4 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.2 | 1.0 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.2 | 1.4 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.2 | 1.5 | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.2 | 2.9 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 0.9 | GO:1903436 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.1 | 1.7 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.1 | 1.8 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 1.1 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 0.8 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 4.8 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 0.9 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 0.5 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.9 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.1 | 3.4 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.1 | 0.9 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.1 | 0.4 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.1 | 1.5 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 4.0 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.1 | 0.8 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.1 | 0.4 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 | 9.3 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.7 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 2.8 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.6 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.0 | 0.4 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 1.0 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.7 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.4 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 6.4 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 2.3 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.7 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 1.8 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 1.2 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 2.1 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.3 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 2.4 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 2.0 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.0 | GO:0051329 | interphase(GO:0051325) mitotic interphase(GO:0051329) mitotic cell cycle phase(GO:0098763) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.9 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.7 | 6.5 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.2 | 3.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.2 | 10.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.2 | 18.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 0.8 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.2 | 5.1 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.2 | 2.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 4.5 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 2.5 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 1.5 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 4.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 7.1 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.0 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 0.5 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 10.1 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 0.4 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.1 | 1.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 2.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.9 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 5.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 5.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 6.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.6 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.2 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.9 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 2.5 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 2.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 2.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 4.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 9.5 | GO:0031252 | cell leading edge(GO:0031252) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.4 | GO:0099530 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 1.1 | 16.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.9 | 6.2 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.8 | 5.5 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.7 | 5.2 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.7 | 4.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.6 | 6.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.5 | 4.5 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.4 | 1.8 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.3 | 2.5 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.3 | 1.2 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.2 | 2.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 0.8 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.2 | 3.7 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 4.4 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.2 | 7.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 0.5 | GO:0033680 | ATP-dependent DNA/RNA helicase activity(GO:0033680) triplex DNA binding(GO:0045142) |
| 0.2 | 5.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.2 | 18.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 3.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 2.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) cobalt ion binding(GO:0050897) |
| 0.1 | 1.0 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 1.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 1.7 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 3.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.4 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 2.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 2.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.4 | GO:0002135 | CTP binding(GO:0002135) |
| 0.0 | 4.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 2.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.9 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 4.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 6.2 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 4.3 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 2.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 5.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.4 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 1.5 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 2.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 1.5 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.1 | GO:0004527 | exonuclease activity(GO:0004527) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 5.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 6.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 4.0 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 2.4 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 2.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.4 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 4.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 11.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 6.2 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.2 | 5.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 3.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 3.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 2.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 2.5 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.9 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.4 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 1.0 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 6.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |