Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
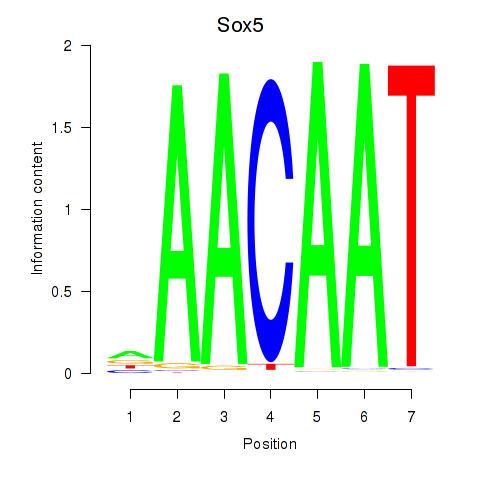
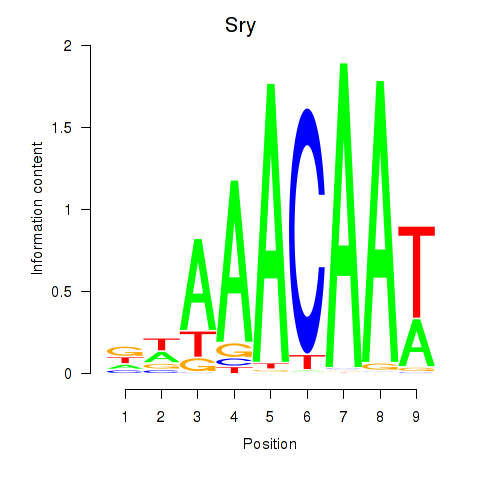
Results for Sox5_Sry
Z-value: 1.23
Transcription factors associated with Sox5_Sry
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox5
|
ENSMUSG00000041540.17 | Sox5 |
|
Sry
|
ENSMUSG00000069036.4 | Sry |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox5 | mm39_v1_chr6_-_144155197_144155294 | 0.38 | 1.1e-03 | Click! |
| Sry | mm39_v1_chrY_-_2663658_2663658 | 0.13 | 2.7e-01 | Click! |
Activity profile of Sox5_Sry motif
Sorted Z-values of Sox5_Sry motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox5_Sry
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.8 | GO:0014728 | regulation of the force of skeletal muscle contraction(GO:0014728) regulation of skeletal muscle contraction by chemo-mechanical energy conversion(GO:0014862) |
| 2.8 | 11.2 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 1.7 | 5.2 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 1.7 | 20.4 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 1.7 | 5.0 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 1.5 | 4.6 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 1.5 | 4.5 | GO:1903465 | motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 1.4 | 5.6 | GO:0072277 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 1.3 | 3.9 | GO:0030824 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 1.3 | 10.3 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 1.2 | 4.9 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 1.2 | 3.7 | GO:0060197 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) cloacal septation(GO:0060197) |
| 1.2 | 3.6 | GO:0090673 | endothelial cell-matrix adhesion(GO:0090673) |
| 1.2 | 4.6 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 1.1 | 4.5 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 1.0 | 6.2 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 1.0 | 5.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 1.0 | 4.1 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 1.0 | 3.1 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 1.0 | 5.9 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 1.0 | 4.0 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 1.0 | 3.0 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 1.0 | 12.6 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 1.0 | 7.6 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.9 | 8.8 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.8 | 4.2 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.7 | 8.8 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.6 | 4.4 | GO:0048840 | otolith development(GO:0048840) |
| 0.6 | 7.9 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.6 | 2.4 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.6 | 2.9 | GO:1902163 | negative regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902163) |
| 0.6 | 2.3 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.6 | 1.7 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.6 | 17.8 | GO:0097435 | fibril organization(GO:0097435) |
| 0.5 | 9.8 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.5 | 2.7 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.5 | 3.2 | GO:0098885 | modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 0.5 | 1.6 | GO:0030070 | insulin processing(GO:0030070) |
| 0.5 | 4.6 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.5 | 1.5 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.5 | 1.5 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.5 | 4.3 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.4 | 1.7 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.4 | 1.2 | GO:0090320 | regulation of chylomicron remnant clearance(GO:0090320) |
| 0.4 | 1.1 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.3 | 3.7 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.3 | 9.0 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.3 | 2.0 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.3 | 1.0 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) negative regulation of histone phosphorylation(GO:0033128) |
| 0.3 | 1.3 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.3 | 1.3 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.3 | 5.7 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.3 | 1.2 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.3 | 2.6 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.3 | 0.9 | GO:1990751 | regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 0.3 | 1.4 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.3 | 2.8 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.3 | 1.4 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.3 | 6.6 | GO:0001553 | luteinization(GO:0001553) |
| 0.3 | 1.3 | GO:0035795 | negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.2 | 2.9 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.2 | 1.9 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.2 | 1.2 | GO:1902608 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.2 | 1.6 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.2 | 1.2 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.2 | 1.6 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.2 | 1.9 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.2 | 2.7 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.2 | 2.7 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 3.4 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.2 | 1.4 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.2 | 1.5 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.2 | 1.2 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.2 | 0.9 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.2 | 1.4 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.2 | 3.2 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.2 | 2.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 1.3 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.2 | 0.8 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.2 | 1.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.2 | 1.4 | GO:0014719 | skeletal muscle satellite cell activation(GO:0014719) negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.2 | 0.9 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) negative regulation of prostatic bud formation(GO:0060686) |
| 0.2 | 1.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.2 | 0.3 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 3.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 0.4 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.1 | 2.6 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 3.4 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.7 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) |
| 0.1 | 0.5 | GO:0045186 | zonula adherens assembly(GO:0045186) |
| 0.1 | 1.5 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 4.7 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.1 | 1.2 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.4 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.1 | 0.5 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.9 | GO:0042938 | antibiotic transport(GO:0042891) dipeptide transport(GO:0042938) |
| 0.1 | 1.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 1.1 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.1 | 1.5 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.4 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 3.6 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 2.8 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 | 2.9 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 | 2.8 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 3.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.6 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.1 | 0.8 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 0.8 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.1 | 3.5 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 3.6 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 0.8 | GO:0042640 | anagen(GO:0042640) |
| 0.1 | 0.7 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.1 | 2.6 | GO:0044247 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 1.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.8 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.6 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.1 | 0.9 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 5.3 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 1.3 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 2.2 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 | 0.6 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 0.7 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 1.1 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.1 | 0.6 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 1.4 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.5 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.1 | 0.8 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 2.6 | GO:0070875 | positive regulation of glycogen biosynthetic process(GO:0045725) positive regulation of glycogen metabolic process(GO:0070875) |
| 0.1 | 0.5 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 1.5 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.6 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 0.5 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.1 | 1.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.2 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.1 | 0.3 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 0.7 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.6 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 1.0 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 0.6 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 0.2 | GO:0021778 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 | 1.0 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 2.1 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 0.9 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.1 | 0.1 | GO:0042253 | granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0042253) regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 1.2 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 1.7 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 1.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 2.3 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.0 | 2.4 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 1.6 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 2.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 1.7 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 1.4 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 1.5 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 9.1 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 0.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 1.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.6 | GO:1901741 | regulation of myoblast fusion(GO:1901739) positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 2.1 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.0 | 0.9 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.6 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 2.1 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 1.3 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.2 | GO:0003170 | heart valve development(GO:0003170) |
| 0.0 | 2.1 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 1.1 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.2 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 5.9 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.5 | GO:0010644 | cell communication by electrical coupling(GO:0010644) |
| 0.0 | 0.3 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.9 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.5 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 2.9 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.5 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 1.7 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 3.9 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 1.5 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 0.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.3 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.0 | 0.4 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 2.9 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.3 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 2.5 | GO:0030168 | platelet activation(GO:0030168) |
| 0.0 | 0.1 | GO:2000292 | negative regulation of eating behavior(GO:1903999) regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.0 | 2.2 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.0 | 2.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 2.4 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.0 | 0.8 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.7 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 0.1 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 | 0.8 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.2 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.3 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 3.1 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.2 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.8 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.0 | 0.0 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.2 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.1 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.7 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 8.9 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 1.0 | 16.7 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.8 | 6.6 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.5 | 11.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.5 | 12.6 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.5 | 3.6 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.4 | 15.9 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.4 | 2.6 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.4 | 10.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.4 | 3.4 | GO:0031673 | H zone(GO:0031673) |
| 0.4 | 5.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.4 | 1.1 | GO:0044317 | rod spherule(GO:0044317) |
| 0.3 | 3.9 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.3 | 0.8 | GO:0005712 | chiasma(GO:0005712) |
| 0.2 | 1.5 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.2 | 1.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 4.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.2 | 1.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 1.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 4.4 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 3.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 3.3 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.2 | 1.6 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.2 | 1.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 1.8 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 4.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 1.3 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.9 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 2.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 1.0 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 5.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 8.7 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 1.7 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 9.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 0.8 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 3.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.9 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 26.0 | GO:0030017 | sarcomere(GO:0030017) |
| 0.1 | 11.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 1.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 1.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 7.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 2.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 2.8 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 17.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.7 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.1 | 2.4 | GO:1990752 | microtubule plus-end(GO:0035371) microtubule end(GO:1990752) |
| 0.1 | 0.3 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.1 | 1.3 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 3.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 1.7 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.1 | 0.5 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.1 | 0.8 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 2.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 2.6 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 1.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 2.6 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 4.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.7 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 1.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 1.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 2.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 2.1 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.8 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.2 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 1.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 12.2 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 1.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.3 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.4 | GO:0005876 | spindle microtubule(GO:0005876) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 16.7 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 1.9 | 5.6 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 1.7 | 13.3 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 1.1 | 4.3 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 1.0 | 15.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.9 | 2.6 | GO:0004135 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.8 | 4.0 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.7 | 11.0 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.7 | 18.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.7 | 4.6 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.6 | 11.2 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.6 | 12.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.6 | 3.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.6 | 1.7 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 0.5 | 1.1 | GO:0002135 | CTP binding(GO:0002135) |
| 0.5 | 10.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.5 | 1.9 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.5 | 2.8 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.4 | 3.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.4 | 4.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.4 | 1.2 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.4 | 1.5 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.3 | 1.7 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.3 | 1.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.3 | 0.9 | GO:0042936 | dipeptide transporter activity(GO:0042936) |
| 0.3 | 2.5 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.2 | 1.2 | GO:0035478 | chylomicron binding(GO:0035478) |
| 0.2 | 1.4 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.2 | 5.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 1.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 0.8 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.2 | 1.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.2 | 1.2 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.2 | 4.6 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.2 | 0.7 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 14.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 1.3 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 4.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 3.7 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 5.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 4.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 1.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 1.7 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 1.3 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.4 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 1.4 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 0.6 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 1.0 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.1 | 4.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.7 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.6 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.1 | 1.5 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.5 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 4.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 3.5 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 0.9 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.5 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 1.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.6 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 1.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 1.5 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 1.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 6.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 4.0 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 3.1 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 2.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 1.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 2.7 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 0.9 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.1 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.3 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 12.9 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.1 | 8.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 0.6 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 2.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 2.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 2.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 2.0 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 2.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 4.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 2.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 1.4 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.0 | 1.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.7 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.4 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 1.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 10.3 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 1.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 3.3 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 9.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 2.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 3.3 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.9 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.3 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 1.0 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 1.3 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.4 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 4.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.5 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 1.7 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 1.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.8 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 3.4 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.9 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 3.9 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 1.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.3 | 16.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.3 | 18.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.2 | 4.0 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.2 | 12.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 3.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 9.6 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.2 | 4.8 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.2 | 1.2 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 8.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 5.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 5.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 7.7 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 18.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 3.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 3.2 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 0.5 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 1.6 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 6.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 1.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 1.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 11.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 0.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 4.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 2.0 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 1.2 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 1.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.4 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 4.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 2.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.6 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 0.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 2.9 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 9.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 6.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.3 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 29.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.4 | 11.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.3 | 3.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 12.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 4.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 6.6 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.2 | 3.0 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 2.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 3.6 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 5.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 2.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 3.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 3.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 1.2 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.1 | 4.5 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 1.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 1.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 6.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 0.7 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 1.5 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 2.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 4.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 4.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 2.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 2.7 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 1.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 1.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 2.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 3.9 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.4 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 4.3 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 9.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.8 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 1.0 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |