Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
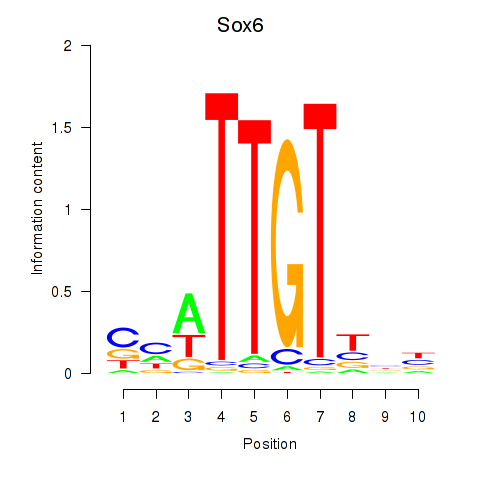
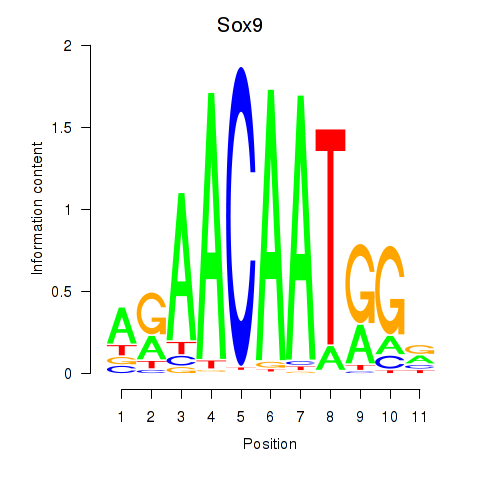
Results for Sox6_Sox9
Z-value: 1.28
Transcription factors associated with Sox6_Sox9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox6
|
ENSMUSG00000051910.14 | Sox6 |
|
Sox9
|
ENSMUSG00000000567.6 | Sox9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox6 | mm39_v1_chr7_-_115459082_115459098 | -0.20 | 8.9e-02 | Click! |
| Sox9 | mm39_v1_chr11_+_112673041_112673059 | 0.12 | 3.0e-01 | Click! |
Activity profile of Sox6_Sox9 motif
Sorted Z-values of Sox6_Sox9 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox6_Sox9
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_119978773 | 26.69 |
ENSMUST00000068698.15
ENSMUST00000215512.2 ENSMUST00000111627.3 ENSMUST00000093773.8 |
Mobp
|
myelin-associated oligodendrocytic basic protein |
| chr9_-_29323032 | 9.69 |
ENSMUST00000115236.2
|
Ntm
|
neurotrimin |
| chr18_+_82572595 | 9.52 |
ENSMUST00000152071.9
ENSMUST00000142850.9 ENSMUST00000080658.12 ENSMUST00000133193.9 ENSMUST00000123251.9 ENSMUST00000153478.9 ENSMUST00000075372.13 ENSMUST00000102812.12 ENSMUST00000062446.15 ENSMUST00000114674.11 ENSMUST00000132369.3 |
Mbp
|
myelin basic protein |
| chr1_-_93029547 | 9.51 |
ENSMUST00000112958.9
ENSMUST00000186861.2 ENSMUST00000171556.8 |
Kif1a
|
kinesin family member 1A |
| chr1_-_93029532 | 9.02 |
ENSMUST00000171796.8
|
Kif1a
|
kinesin family member 1A |
| chr14_-_52151537 | 8.71 |
ENSMUST00000227402.2
ENSMUST00000227237.2 |
Ndrg2
|
N-myc downstream regulated gene 2 |
| chr1_-_46927230 | 8.45 |
ENSMUST00000185520.2
|
Slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr15_-_37458768 | 7.91 |
ENSMUST00000116445.9
|
Ncald
|
neurocalcin delta |
| chr5_+_27109679 | 7.85 |
ENSMUST00000120555.8
|
Dpp6
|
dipeptidylpeptidase 6 |
| chr9_-_29323500 | 7.84 |
ENSMUST00000115237.8
|
Ntm
|
neurotrimin |
| chr18_-_43192483 | 7.79 |
ENSMUST00000025377.14
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr10_+_29019645 | 7.73 |
ENSMUST00000092629.4
|
Soga3
|
SOGA family member 3 |
| chr1_-_93029576 | 7.37 |
ENSMUST00000190723.7
|
Kif1a
|
kinesin family member 1A |
| chr13_+_43276323 | 6.75 |
ENSMUST00000136576.8
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr4_+_140970161 | 6.44 |
ENSMUST00000138096.8
ENSMUST00000006618.9 ENSMUST00000125392.8 |
Arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr15_-_90934059 | 6.40 |
ENSMUST00000109288.9
ENSMUST00000100304.11 |
Kif21a
|
kinesin family member 21A |
| chr5_-_146521629 | 6.11 |
ENSMUST00000200112.2
ENSMUST00000197431.2 ENSMUST00000197825.2 |
Gpr12
|
G-protein coupled receptor 12 |
| chr11_-_68848271 | 5.99 |
ENSMUST00000108671.2
|
Arhgef15
|
Rho guanine nucleotide exchange factor (GEF) 15 |
| chr9_-_75504926 | 5.90 |
ENSMUST00000164100.2
|
Tmod2
|
tropomodulin 2 |
| chr19_+_44282113 | 5.87 |
ENSMUST00000026221.7
|
Scd2
|
stearoyl-Coenzyme A desaturase 2 |
| chr13_+_88969591 | 5.64 |
ENSMUST00000118731.8
ENSMUST00000081769.13 |
Edil3
|
EGF-like repeats and discoidin I-like domains 3 |
| chr14_+_66581818 | 5.61 |
ENSMUST00000118426.8
ENSMUST00000121955.8 ENSMUST00000120229.8 ENSMUST00000134440.2 |
Stmn4
|
stathmin-like 4 |
| chrX_-_23151771 | 5.48 |
ENSMUST00000115319.9
|
Klhl13
|
kelch-like 13 |
| chr5_+_67125759 | 5.15 |
ENSMUST00000238993.2
ENSMUST00000038188.14 |
Limch1
|
LIM and calponin homology domains 1 |
| chr14_+_66581745 | 4.88 |
ENSMUST00000152093.8
ENSMUST00000074523.13 |
Stmn4
|
stathmin-like 4 |
| chr8_+_45960931 | 4.85 |
ENSMUST00000171337.10
ENSMUST00000067107.15 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr13_+_88969725 | 4.82 |
ENSMUST00000043111.7
|
Edil3
|
EGF-like repeats and discoidin I-like domains 3 |
| chr3_+_20011405 | 4.81 |
ENSMUST00000108325.9
|
Cp
|
ceruloplasmin |
| chr11_-_98220466 | 4.78 |
ENSMUST00000041685.7
|
Neurod2
|
neurogenic differentiation 2 |
| chr18_-_23171713 | 4.63 |
ENSMUST00000081423.13
|
Nol4
|
nucleolar protein 4 |
| chr12_+_29578354 | 4.62 |
ENSMUST00000218583.2
ENSMUST00000049784.17 |
Myt1l
|
myelin transcription factor 1-like |
| chr12_+_119909692 | 4.61 |
ENSMUST00000058644.15
|
Tmem196
|
transmembrane protein 196 |
| chr3_+_20011201 | 4.60 |
ENSMUST00000091309.12
ENSMUST00000108329.8 ENSMUST00000003714.13 |
Cp
|
ceruloplasmin |
| chr5_+_34140777 | 4.59 |
ENSMUST00000094869.12
ENSMUST00000114383.8 |
Gm1673
|
predicted gene 1673 |
| chr8_+_45960804 | 4.52 |
ENSMUST00000067065.14
ENSMUST00000124544.8 ENSMUST00000138049.9 ENSMUST00000132139.9 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr2_-_73605684 | 4.45 |
ENSMUST00000112024.10
ENSMUST00000180045.8 |
Chn1
|
chimerin 1 |
| chr2_+_102488985 | 4.31 |
ENSMUST00000080210.10
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr17_-_24863907 | 4.21 |
ENSMUST00000234505.2
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr4_+_48585275 | 4.19 |
ENSMUST00000123476.8
|
Tmeff1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr15_+_6416229 | 4.19 |
ENSMUST00000110664.9
ENSMUST00000110663.9 ENSMUST00000161812.8 ENSMUST00000160134.8 |
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr4_+_48585135 | 4.15 |
ENSMUST00000030032.13
|
Tmeff1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr18_+_37813286 | 4.15 |
ENSMUST00000192931.2
|
Pcdhgb1
|
protocadherin gamma subfamily B, 1 |
| chr9_+_108924457 | 4.10 |
ENSMUST00000072093.13
|
Plxnb1
|
plexin B1 |
| chr17_-_67354971 | 4.08 |
ENSMUST00000224862.2
|
Ptprm
|
protein tyrosine phosphatase, receptor type, M |
| chr6_+_82018604 | 4.05 |
ENSMUST00000042974.15
|
Eva1a
|
eva-1 homolog A (C. elegans) |
| chr10_-_30531832 | 4.00 |
ENSMUST00000217138.2
ENSMUST00000217644.2 ENSMUST00000216172.2 |
Ncoa7
|
nuclear receptor coactivator 7 |
| chr1_-_16589511 | 3.98 |
ENSMUST00000162751.8
ENSMUST00000027052.13 ENSMUST00000149320.9 |
Stau2
|
staufen double-stranded RNA binding protein 2 |
| chr15_+_6416079 | 3.97 |
ENSMUST00000080880.12
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr3_-_116762476 | 3.96 |
ENSMUST00000119557.8
|
Palmd
|
palmdelphin |
| chr1_+_158189831 | 3.94 |
ENSMUST00000193042.6
ENSMUST00000046110.16 |
Astn1
|
astrotactin 1 |
| chr17_-_24863956 | 3.93 |
ENSMUST00000019684.13
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr6_-_42686970 | 3.91 |
ENSMUST00000045054.11
|
Tcaf1
|
TRPM8 channel-associated factor 1 |
| chr13_-_93774469 | 3.85 |
ENSMUST00000099309.6
|
Bhmt
|
betaine-homocysteine methyltransferase |
| chr3_+_20011251 | 3.82 |
ENSMUST00000108328.8
|
Cp
|
ceruloplasmin |
| chr11_+_67477501 | 3.79 |
ENSMUST00000108680.2
|
Gas7
|
growth arrest specific 7 |
| chr1_+_17215581 | 3.74 |
ENSMUST00000026879.8
|
Gdap1
|
ganglioside-induced differentiation-associated-protein 1 |
| chr12_+_29988035 | 3.68 |
ENSMUST00000122328.8
ENSMUST00000118321.3 |
Pxdn
|
peroxidasin |
| chr17_-_90395771 | 3.66 |
ENSMUST00000197268.5
ENSMUST00000173917.8 |
Nrxn1
|
neurexin I |
| chr6_-_128558560 | 3.66 |
ENSMUST00000060574.9
|
A2ml1
|
alpha-2-macroglobulin like 1 |
| chr7_+_141503719 | 3.65 |
ENSMUST00000105989.9
ENSMUST00000075528.12 ENSMUST00000174499.8 |
Brsk2
|
BR serine/threonine kinase 2 |
| chr18_-_16942289 | 3.65 |
ENSMUST00000025166.14
|
Cdh2
|
cadherin 2 |
| chr9_-_64928927 | 3.53 |
ENSMUST00000036615.7
|
Hacd3
|
3-hydroxyacyl-CoA dehydratase 3 |
| chr8_+_46081213 | 3.52 |
ENSMUST00000130850.8
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr18_+_37884653 | 3.50 |
ENSMUST00000194928.2
|
Pcdhgb7
|
protocadherin gamma subfamily B, 7 |
| chr12_+_40495951 | 3.50 |
ENSMUST00000037488.8
|
Dock4
|
dedicator of cytokinesis 4 |
| chr2_-_101713355 | 3.48 |
ENSMUST00000154525.2
|
Prr5l
|
proline rich 5 like |
| chr10_-_30531768 | 3.47 |
ENSMUST00000092610.12
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chr13_-_4200627 | 3.46 |
ENSMUST00000110704.9
ENSMUST00000021635.9 |
Akr1c18
|
aldo-keto reductase family 1, member C18 |
| chr11_+_67477347 | 3.45 |
ENSMUST00000108682.9
|
Gas7
|
growth arrest specific 7 |
| chr14_-_70855980 | 3.44 |
ENSMUST00000228001.2
|
Dmtn
|
dematin actin binding protein |
| chr5_+_142946598 | 3.43 |
ENSMUST00000129306.4
|
Fscn1
|
fascin actin-bundling protein 1 |
| chr12_-_72283465 | 3.43 |
ENSMUST00000021497.16
ENSMUST00000137990.2 |
Rtn1
|
reticulon 1 |
| chr18_+_37622518 | 3.42 |
ENSMUST00000055949.4
|
Pcdhb18
|
protocadherin beta 18 |
| chr18_+_37610858 | 3.42 |
ENSMUST00000051442.7
|
Pcdhb16
|
protocadherin beta 16 |
| chr1_+_153541020 | 3.40 |
ENSMUST00000152114.8
ENSMUST00000111812.8 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr15_-_88863210 | 3.40 |
ENSMUST00000042594.13
ENSMUST00000109368.2 |
Mlc1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 homolog (human) |
| chr1_+_146373352 | 3.39 |
ENSMUST00000132847.8
ENSMUST00000166814.8 |
Brinp3
|
bone morphogenetic protein/retinoic acid inducible neural specific 3 |
| chr18_+_37875135 | 3.37 |
ENSMUST00000003599.9
|
Pcdhgb6
|
protocadherin gamma subfamily B, 6 |
| chr5_+_14564932 | 3.31 |
ENSMUST00000182407.8
ENSMUST00000030691.17 |
Pclo
|
piccolo (presynaptic cytomatrix protein) |
| chr9_+_27702243 | 3.27 |
ENSMUST00000115243.9
|
Opcml
|
opioid binding protein/cell adhesion molecule-like |
| chr6_-_12749192 | 3.26 |
ENSMUST00000172356.8
|
Thsd7a
|
thrombospondin, type I, domain containing 7A |
| chr4_+_42158092 | 3.26 |
ENSMUST00000098122.3
|
Gm13306
|
predicted gene 13306 |
| chr2_-_170248421 | 3.24 |
ENSMUST00000154650.8
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr11_-_114686712 | 3.20 |
ENSMUST00000000206.4
|
Btbd17
|
BTB (POZ) domain containing 17 |
| chr4_-_114991478 | 3.18 |
ENSMUST00000106545.8
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr5_+_142946098 | 3.17 |
ENSMUST00000031565.15
ENSMUST00000198017.5 |
Fscn1
|
fascin actin-bundling protein 1 |
| chr14_+_5894220 | 3.17 |
ENSMUST00000063750.8
|
Rarb
|
retinoic acid receptor, beta |
| chr1_+_153776323 | 3.16 |
ENSMUST00000140685.4
ENSMUST00000139476.8 |
Glul
|
glutamate-ammonia ligase (glutamine synthetase) |
| chr2_+_156262756 | 3.14 |
ENSMUST00000103137.10
|
Epb41l1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr2_-_73605387 | 3.12 |
ENSMUST00000166199.9
|
Chn1
|
chimerin 1 |
| chr6_-_12749409 | 3.10 |
ENSMUST00000119581.7
|
Thsd7a
|
thrombospondin, type I, domain containing 7A |
| chr18_+_37440497 | 3.10 |
ENSMUST00000056712.4
|
Pcdhb4
|
protocadherin beta 4 |
| chr8_-_4309257 | 3.10 |
ENSMUST00000053252.9
|
Ctxn1
|
cortexin 1 |
| chr5_+_34140877 | 3.09 |
ENSMUST00000114382.2
|
Gm1673
|
predicted gene 1673 |
| chr5_-_8417982 | 3.05 |
ENSMUST00000088761.11
ENSMUST00000115386.8 ENSMUST00000050166.14 ENSMUST00000046838.14 ENSMUST00000115388.9 ENSMUST00000088744.12 ENSMUST00000115385.2 |
Adam22
|
a disintegrin and metallopeptidase domain 22 |
| chr6_-_124718316 | 3.03 |
ENSMUST00000004389.6
|
Grcc10
|
gene rich cluster, C10 gene |
| chr4_+_94627755 | 3.03 |
ENSMUST00000071168.6
|
Tek
|
TEK receptor tyrosine kinase |
| chr6_-_122317484 | 3.00 |
ENSMUST00000112600.9
|
Phc1
|
polyhomeotic 1 |
| chr11_+_67345895 | 2.97 |
ENSMUST00000108681.9
|
Gas7
|
growth arrest specific 7 |
| chr2_+_3115250 | 2.94 |
ENSMUST00000072955.12
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr3_-_123484499 | 2.94 |
ENSMUST00000154668.8
|
Ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr1_+_158190090 | 2.91 |
ENSMUST00000194369.6
ENSMUST00000195311.6 |
Astn1
|
astrotactin 1 |
| chr2_+_102489558 | 2.89 |
ENSMUST00000111213.8
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr7_+_128290204 | 2.86 |
ENSMUST00000118605.2
|
Inpp5f
|
inositol polyphosphate-5-phosphatase F |
| chr2_+_31135813 | 2.84 |
ENSMUST00000000199.8
|
Ncs1
|
neuronal calcium sensor 1 |
| chrX_-_166638057 | 2.80 |
ENSMUST00000238211.2
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr1_-_168259264 | 2.80 |
ENSMUST00000176790.8
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr11_+_87651359 | 2.79 |
ENSMUST00000039627.12
ENSMUST00000100644.10 |
Tspoap1
|
TSPO associated protein 1 |
| chr13_+_93441447 | 2.79 |
ENSMUST00000109497.8
ENSMUST00000109498.8 ENSMUST00000060490.11 ENSMUST00000109492.9 ENSMUST00000109496.8 ENSMUST00000109495.8 |
Homer1
|
homer scaffolding protein 1 |
| chr6_+_22875494 | 2.79 |
ENSMUST00000090568.7
|
Ptprz1
|
protein tyrosine phosphatase, receptor type Z, polypeptide 1 |
| chr3_-_116762617 | 2.77 |
ENSMUST00000143611.2
ENSMUST00000040097.14 |
Palmd
|
palmdelphin |
| chr2_-_180596469 | 2.77 |
ENSMUST00000148905.8
ENSMUST00000103053.10 ENSMUST00000108873.9 |
Nkain4
|
Na+/K+ transporting ATPase interacting 4 |
| chr3_+_94385602 | 2.74 |
ENSMUST00000199884.5
ENSMUST00000198316.5 ENSMUST00000197558.5 |
Celf3
|
CUGBP, Elav-like family member 3 |
| chr1_-_168259070 | 2.72 |
ENSMUST00000064438.11
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr4_+_43406435 | 2.71 |
ENSMUST00000098106.9
ENSMUST00000139198.2 |
Rusc2
|
RUN and SH3 domain containing 2 |
| chr11_-_11840367 | 2.70 |
ENSMUST00000155690.2
|
Ddc
|
dopa decarboxylase |
| chr5_-_132570710 | 2.70 |
ENSMUST00000182974.9
|
Auts2
|
autism susceptibility candidate 2 |
| chr2_+_158508609 | 2.70 |
ENSMUST00000103116.10
|
Ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr11_-_11840394 | 2.68 |
ENSMUST00000109659.9
|
Ddc
|
dopa decarboxylase |
| chr9_+_13677266 | 2.68 |
ENSMUST00000152532.8
|
Mtmr2
|
myotubularin related protein 2 |
| chr1_-_16590244 | 2.63 |
ENSMUST00000144138.4
ENSMUST00000145092.8 ENSMUST00000131257.9 ENSMUST00000153966.9 ENSMUST00000162435.8 |
Stau2
|
staufen double-stranded RNA binding protein 2 |
| chr6_-_23839136 | 2.62 |
ENSMUST00000166458.9
ENSMUST00000142913.9 ENSMUST00000069074.14 ENSMUST00000115361.9 ENSMUST00000018122.14 ENSMUST00000115356.3 |
Cadps2
|
Ca2+-dependent activator protein for secretion 2 |
| chr11_-_59073635 | 2.59 |
ENSMUST00000108793.9
|
Gjc2
|
gap junction protein, gamma 2 |
| chr13_+_93441307 | 2.57 |
ENSMUST00000080127.12
|
Homer1
|
homer scaffolding protein 1 |
| chr17_-_10501816 | 2.50 |
ENSMUST00000233684.2
|
Qk
|
quaking, KH domain containing RNA binding |
| chr8_+_22996233 | 2.49 |
ENSMUST00000210854.2
|
Slc20a2
|
solute carrier family 20, member 2 |
| chr6_-_13839914 | 2.46 |
ENSMUST00000060442.14
|
Gpr85
|
G protein-coupled receptor 85 |
| chr6_-_31540913 | 2.46 |
ENSMUST00000026698.8
|
Podxl
|
podocalyxin-like |
| chr12_+_69343450 | 2.46 |
ENSMUST00000021362.5
|
Klhdc2
|
kelch domain containing 2 |
| chr7_+_141503583 | 2.45 |
ENSMUST00000172652.8
|
Brsk2
|
BR serine/threonine kinase 2 |
| chr10_+_90665270 | 2.44 |
ENSMUST00000182202.8
ENSMUST00000182966.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr16_-_59452883 | 2.42 |
ENSMUST00000118438.8
|
Arl6
|
ADP-ribosylation factor-like 6 |
| chr8_-_71848429 | 2.40 |
ENSMUST00000049184.9
|
Ushbp1
|
USH1 protein network component harmonin binding protein 1 |
| chr17_+_24026892 | 2.39 |
ENSMUST00000191385.3
|
Srrm2
|
serine/arginine repetitive matrix 2 |
| chr18_+_37858753 | 2.39 |
ENSMUST00000066149.9
|
Pcdhga8
|
protocadherin gamma subfamily A, 8 |
| chr8_+_26008799 | 2.34 |
ENSMUST00000119398.10
ENSMUST00000117179.9 |
Fgfr1
|
fibroblast growth factor receptor 1 |
| chr11_-_102338473 | 2.34 |
ENSMUST00000049057.5
|
Fam171a2
|
family with sequence similarity 171, member A2 |
| chr11_-_3813895 | 2.32 |
ENSMUST00000070552.14
|
Osbp2
|
oxysterol binding protein 2 |
| chr2_+_67948057 | 2.30 |
ENSMUST00000112346.3
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr3_+_94385661 | 2.27 |
ENSMUST00000200342.5
|
Celf3
|
CUGBP, Elav-like family member 3 |
| chr12_-_58315949 | 2.27 |
ENSMUST00000062254.4
|
Clec14a
|
C-type lectin domain family 14, member a |
| chr12_+_117652526 | 2.23 |
ENSMUST00000222185.2
|
Rapgef5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr4_+_94627513 | 2.23 |
ENSMUST00000073939.13
ENSMUST00000102798.8 |
Tek
|
TEK receptor tyrosine kinase |
| chr8_-_9821021 | 2.23 |
ENSMUST00000208933.2
ENSMUST00000110969.5 |
Fam155a
|
family with sequence similarity 155, member A |
| chr5_+_67125902 | 2.23 |
ENSMUST00000127184.8
|
Limch1
|
LIM and calponin homology domains 1 |
| chr2_+_67935015 | 2.22 |
ENSMUST00000042456.4
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr19_-_41065544 | 2.19 |
ENSMUST00000087176.8
|
Opalin
|
oligodendrocytic myelin paranodal and inner loop protein |
| chr17_-_90395568 | 2.19 |
ENSMUST00000173222.2
|
Nrxn1
|
neurexin I |
| chr2_-_180596413 | 2.19 |
ENSMUST00000139929.8
|
Nkain4
|
Na+/K+ transporting ATPase interacting 4 |
| chr17_+_55752485 | 2.18 |
ENSMUST00000025000.4
|
St6gal2
|
beta galactoside alpha 2,6 sialyltransferase 2 |
| chr10_+_90665399 | 2.17 |
ENSMUST00000179694.9
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr15_+_41694317 | 2.17 |
ENSMUST00000166917.3
ENSMUST00000230127.2 ENSMUST00000230131.2 |
Oxr1
|
oxidation resistance 1 |
| chr18_+_37143758 | 2.16 |
ENSMUST00000115657.10
ENSMUST00000192447.6 |
Pcdha11
|
protocadherin alpha 11 |
| chrX_-_16777913 | 2.15 |
ENSMUST00000040134.8
|
Ndp
|
Norrie disease (pseudoglioma) (human) |
| chr1_+_158189900 | 2.14 |
ENSMUST00000170718.7
|
Astn1
|
astrotactin 1 |
| chr9_+_72832904 | 2.11 |
ENSMUST00000038489.6
|
Pygo1
|
pygopus 1 |
| chrX_+_99811325 | 2.10 |
ENSMUST00000000901.13
ENSMUST00000113736.9 ENSMUST00000087984.11 |
Dlg3
|
discs large MAGUK scaffold protein 3 |
| chr18_+_37853415 | 2.06 |
ENSMUST00000195363.2
|
Pcdhgb4
|
protocadherin gamma subfamily B, 4 |
| chr8_+_26008773 | 2.06 |
ENSMUST00000084027.13
ENSMUST00000178276.8 ENSMUST00000179592.8 |
Fgfr1
|
fibroblast growth factor receptor 1 |
| chr5_-_131645437 | 2.04 |
ENSMUST00000161804.9
|
Auts2
|
autism susceptibility candidate 2 |
| chr11_+_93935021 | 2.04 |
ENSMUST00000075695.13
ENSMUST00000092777.11 |
Spag9
|
sperm associated antigen 9 |
| chr18_+_37617848 | 2.03 |
ENSMUST00000053856.6
|
Pcdhb17
|
protocadherin beta 17 |
| chrX_-_101114906 | 2.02 |
ENSMUST00000188731.2
|
Rtl5
|
retrotransposon Gag like 5 |
| chr3_-_87934772 | 2.01 |
ENSMUST00000005014.9
|
Hapln2
|
hyaluronan and proteoglycan link protein 2 |
| chr11_+_29642937 | 2.00 |
ENSMUST00000102843.10
ENSMUST00000102842.10 ENSMUST00000078830.11 ENSMUST00000170731.8 |
Rtn4
|
reticulon 4 |
| chr7_-_142220553 | 1.99 |
ENSMUST00000105935.8
|
Igf2
|
insulin-like growth factor 2 |
| chr5_+_75316552 | 1.95 |
ENSMUST00000168162.5
|
Pdgfra
|
platelet derived growth factor receptor, alpha polypeptide |
| chr11_-_31621863 | 1.93 |
ENSMUST00000058060.14
|
Bod1
|
biorientation of chromosomes in cell division 1 |
| chr18_+_37554471 | 1.93 |
ENSMUST00000053073.6
|
Pcdhb11
|
protocadherin beta 11 |
| chr18_+_37466877 | 1.89 |
ENSMUST00000194655.2
ENSMUST00000061717.4 |
Pcdhb6
|
protocadherin beta 6 |
| chr4_-_63072367 | 1.87 |
ENSMUST00000030041.5
|
Ambp
|
alpha 1 microglobulin/bikunin precursor |
| chr18_+_37864045 | 1.83 |
ENSMUST00000192535.2
|
Pcdhgb5
|
protocadherin gamma subfamily B, 5 |
| chr11_-_49077864 | 1.83 |
ENSMUST00000153999.3
ENSMUST00000066531.13 |
Btnl9
|
butyrophilin-like 9 |
| chrX_+_92698469 | 1.82 |
ENSMUST00000113933.9
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr7_-_126275529 | 1.82 |
ENSMUST00000106372.11
ENSMUST00000155419.3 ENSMUST00000106373.9 |
Sult1a1
|
sulfotransferase family 1A, phenol-preferring, member 1 |
| chr4_+_42655251 | 1.82 |
ENSMUST00000177785.3
|
Ccl27b
|
chemokine (C-C motif) ligand 27b |
| chr2_-_65397850 | 1.80 |
ENSMUST00000238483.2
ENSMUST00000100069.9 |
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr7_+_143792455 | 1.77 |
ENSMUST00000239495.2
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr1_-_135302971 | 1.77 |
ENSMUST00000041240.4
|
Shisa4
|
shisa family member 4 |
| chr12_+_52746158 | 1.76 |
ENSMUST00000095737.5
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr12_+_31315270 | 1.75 |
ENSMUST00000002979.16
ENSMUST00000239496.2 ENSMUST00000170495.3 |
Lamb1
|
laminin B1 |
| chr5_-_123320767 | 1.74 |
ENSMUST00000154713.8
ENSMUST00000031398.14 |
Hpd
|
4-hydroxyphenylpyruvic acid dioxygenase |
| chr5_+_65288418 | 1.73 |
ENSMUST00000101191.10
ENSMUST00000204348.3 |
Klhl5
|
kelch-like 5 |
| chr11_+_93935066 | 1.71 |
ENSMUST00000103168.10
|
Spag9
|
sperm associated antigen 9 |
| chr19_-_8382424 | 1.68 |
ENSMUST00000064507.12
ENSMUST00000120540.2 ENSMUST00000096269.11 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr8_+_45960855 | 1.67 |
ENSMUST00000141039.8
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr14_-_63415235 | 1.66 |
ENSMUST00000054963.10
|
Fdft1
|
farnesyl diphosphate farnesyl transferase 1 |
| chr8_-_69636825 | 1.64 |
ENSMUST00000185176.8
|
Lzts1
|
leucine zipper, putative tumor suppressor 1 |
| chrX_-_135769285 | 1.63 |
ENSMUST00000058814.7
|
Rab9b
|
RAB9B, member RAS oncogene family |
| chr18_+_37880027 | 1.62 |
ENSMUST00000193404.2
|
Pcdhga10
|
protocadherin gamma subfamily A, 10 |
| chr14_-_20668406 | 1.60 |
ENSMUST00000035340.14
|
Usp54
|
ubiquitin specific peptidase 54 |
| chr5_-_148329615 | 1.58 |
ENSMUST00000138257.8
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr4_-_63073028 | 1.57 |
ENSMUST00000142901.2
|
Ambp
|
alpha 1 microglobulin/bikunin precursor |
| chr18_+_37630044 | 1.57 |
ENSMUST00000059571.7
|
Pcdhb19
|
protocadherin beta 19 |
| chr8_-_106015682 | 1.56 |
ENSMUST00000212922.2
ENSMUST00000212219.2 |
4931428F04Rik
|
RIKEN cDNA 4931428F04 gene |
| chr9_+_122752116 | 1.54 |
ENSMUST00000051667.14
|
Zfp105
|
zinc finger protein 105 |
| chrX_+_111510223 | 1.54 |
ENSMUST00000113409.8
|
Zfp711
|
zinc finger protein 711 |
| chr14_-_28691423 | 1.53 |
ENSMUST00000225985.2
|
Cacna2d3
|
calcium channel, voltage-dependent, alpha2/delta subunit 3 |
| chr3_+_135531548 | 1.48 |
ENSMUST00000167390.8
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chrM_+_7758 | 1.47 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.8 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 2.4 | 9.5 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 2.2 | 26.7 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 2.0 | 8.0 | GO:0098961 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 1.5 | 4.4 | GO:0021837 | motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 1.5 | 8.7 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 1.4 | 8.2 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 1.2 | 4.7 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 1.2 | 7.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 1.2 | 3.5 | GO:0009753 | sesquiterpenoid metabolic process(GO:0006714) response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 1.1 | 3.3 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 1.1 | 25.9 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 1.1 | 5.3 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 1.0 | 7.2 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 1.0 | 4.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 1.0 | 5.9 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.8 | 3.3 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 0.8 | 5.4 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.8 | 8.5 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.7 | 9.0 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.7 | 6.6 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.7 | 2.9 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.7 | 4.9 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.7 | 3.5 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.7 | 2.7 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.7 | 5.3 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.6 | 3.9 | GO:0043102 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.6 | 2.6 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.6 | 3.2 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.6 | 3.0 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.6 | 3.0 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.6 | 3.5 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.6 | 1.8 | GO:0090320 | regulation of chylomicron remnant clearance(GO:0090320) |
| 0.6 | 3.4 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.6 | 1.7 | GO:0044108 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) cellular alcohol metabolic process(GO:0044107) cellular alcohol biosynthetic process(GO:0044108) |
| 0.5 | 4.6 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.5 | 9.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.5 | 4.5 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.5 | 2.0 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.5 | 3.9 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.5 | 2.4 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.5 | 1.4 | GO:0035602 | orbitofrontal cortex development(GO:0021769) fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) fibroblast growth factor receptor signaling pathway involved in orbitofrontal cortex development(GO:0035607) coronal suture morphogenesis(GO:0060365) |
| 0.5 | 5.9 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.4 | 7.8 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.4 | 1.3 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.4 | 4.6 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.4 | 1.2 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.4 | 3.7 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.4 | 2.9 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.4 | 2.0 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.4 | 2.4 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.4 | 3.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.4 | 1.9 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.4 | 14.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.4 | 6.5 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.3 | 1.0 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.3 | 3.4 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.3 | 2.6 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.3 | 3.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.3 | 2.8 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.3 | 0.9 | GO:0090673 | endothelial cell-matrix adhesion(GO:0090673) |
| 0.3 | 0.9 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.3 | 2.7 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.3 | 1.8 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.3 | 1.1 | GO:0038016 | insulin receptor internalization(GO:0038016) |
| 0.3 | 1.6 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.3 | 2.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.3 | 3.4 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.3 | 0.8 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.3 | 7.1 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.2 | 1.2 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.2 | 3.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.2 | 2.5 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.2 | 2.0 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 | 2.0 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.2 | 14.6 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 2.7 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.2 | 1.0 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.2 | 1.7 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 0.7 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.2 | 7.6 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.2 | 5.3 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.2 | 1.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 2.2 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 2.2 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 11.2 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.4 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.1 | 3.1 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 0.6 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 2.5 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 7.8 | GO:1901381 | positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.1 | 0.4 | GO:1903760 | regulation of potassium ion import(GO:1903286) regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.1 | 3.7 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 1.7 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 9.0 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 3.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.8 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.1 | 0.6 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.1 | 3.2 | GO:0005980 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 0.2 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 1.1 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.1 | 1.0 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 1.7 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 4.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 1.0 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 3.7 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 0.4 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 1.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 1.6 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 0.4 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 | 1.8 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 2.5 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.1 | 0.8 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 0.3 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.1 | 0.9 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 | 0.3 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.1 | 2.4 | GO:0010644 | cell communication by electrical coupling(GO:0010644) |
| 0.1 | 3.2 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.1 | 4.8 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 2.7 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 2.5 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.1 | 1.4 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.1 | 0.7 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 14.3 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.1 | 9.7 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.1 | 0.4 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 7.9 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.1 | 0.7 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.4 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 | 10.1 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.1 | 2.2 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 1.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.6 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.1 | 1.7 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 0.3 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.6 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 0.6 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 1.1 | GO:2001273 | regulation of glucose import in response to insulin stimulus(GO:2001273) |
| 0.1 | 1.3 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.1 | 0.9 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.1 | 0.7 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 1.8 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.9 | GO:0021895 | cerebral cortex neuron differentiation(GO:0021895) |
| 0.0 | 0.5 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 2.2 | GO:0002042 | cell migration involved in sprouting angiogenesis(GO:0002042) |
| 0.0 | 0.6 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 1.6 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.1 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.0 | 6.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 1.4 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 0.5 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 1.2 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 1.0 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.3 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 5.1 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 0.2 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 3.2 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 0.4 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.0 | 0.1 | GO:0098909 | membrane depolarization during SA node cell action potential(GO:0086046) regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.0 | 6.4 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 6.1 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 1.1 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 2.0 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 5.9 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.9 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 1.6 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.2 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.7 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.7 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.4 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.0 | 0.3 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.1 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 1.6 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.3 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 8.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 1.3 | 6.6 | GO:0044393 | microspike(GO:0044393) |
| 1.2 | 9.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 1.1 | 3.3 | GO:0044317 | rod spherule(GO:0044317) |
| 1.0 | 2.9 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.7 | 2.8 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.5 | 3.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.4 | 1.8 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.4 | 8.3 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.4 | 32.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.3 | 3.9 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.3 | 8.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 0.7 | GO:0097632 | extrinsic component of pre-autophagosomal structure membrane(GO:0097632) |
| 0.2 | 7.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 3.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.2 | 2.6 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.2 | 27.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.2 | 1.4 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.2 | 1.2 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.2 | 1.0 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.2 | 2.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 2.7 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.2 | 19.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 9.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 10.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 0.6 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 4.5 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 1.8 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 0.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.1 | 3.7 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 3.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 17.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.9 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 3.2 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.1 | 3.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 4.0 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 0.5 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 1.7 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 1.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.6 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 3.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 13.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 11.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 3.2 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 6.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 2.2 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.1 | 2.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.4 | GO:0043219 | lateral loop(GO:0043219) |
| 0.1 | 0.9 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 0.3 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.1 | 2.0 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 0.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 1.0 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 11.5 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 0.6 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 1.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.4 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 1.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 11.6 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 6.4 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.9 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 2.5 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 4.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.8 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.6 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 4.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 3.4 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 5.2 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 9.9 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 7.0 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 1.6 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 4.5 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 2.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 19.7 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 25.5 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 0.6 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 1.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 1.3 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 7.1 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 0.8 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 38.7 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.3 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.6 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.6 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 36.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 1.3 | 5.4 | GO:0036468 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 1.2 | 7.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 1.2 | 14.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 1.2 | 7.1 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 1.2 | 3.5 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 1.1 | 4.5 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.8 | 2.5 | GO:0004133 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.7 | 2.2 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.7 | 3.5 | GO:0102345 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.7 | 2.8 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.7 | 4.6 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.7 | 2.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.6 | 1.9 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 0.6 | 1.8 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.6 | 1.7 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) quercetin 2,3-dioxygenase activity(GO:0008127) |
| 0.6 | 2.9 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.6 | 3.4 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.5 | 2.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.5 | 5.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.5 | 6.5 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.5 | 2.5 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.5 | 3.9 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.5 | 1.8 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.4 | 1.8 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.4 | 8.2 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.4 | 5.9 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.4 | 5.0 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.4 | 5.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.4 | 1.8 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.4 | 1.8 | GO:0035478 | chylomicron binding(GO:0035478) |
| 0.3 | 1.7 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.3 | 2.6 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.3 | 2.9 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.3 | 1.3 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.3 | 4.1 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.3 | 1.2 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.3 | 6.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.3 | 5.3 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.3 | 31.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.3 | 6.1 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.3 | 4.6 | GO:0050897 | retinoic acid-responsive element binding(GO:0044323) cobalt ion binding(GO:0050897) |
| 0.3 | 1.4 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.3 | 2.0 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.3 | 10.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 3.9 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 3.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 2.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 3.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.2 | 0.9 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.2 | 0.9 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 0.2 | 0.8 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.2 | 12.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 4.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 8.1 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.2 | 1.6 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.2 | 0.7 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.2 | 2.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.2 | 0.7 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.2 | 7.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 7.3 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.2 | 1.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 2.6 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 1.0 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 15.1 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 13.1 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 0.6 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.1 | 1.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.2 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 1.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 1.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 1.4 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 2.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 1.7 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 1.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 7.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 3.2 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 4.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.4 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 3.1 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.1 | 0.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 2.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.3 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 0.1 | 2.0 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 10.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 3.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.7 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 3.3 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 3.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.7 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 1.0 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 8.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.3 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.2 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.8 | GO:0004549 | tRNA-specific ribonuclease activity(GO:0004549) |
| 0.0 | 2.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 6.7 | GO:0008144 | drug binding(GO:0008144) |
| 0.0 | 1.0 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.9 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.3 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 1.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 6.1 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 2.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.9 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 3.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 2.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 11.4 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 3.4 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.3 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.3 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 3.7 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 1.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 3.4 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 1.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.6 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 2.8 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.0 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.6 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 10.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 2.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 6.8 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 2.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 7.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 6.0 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 2.8 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 4.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 3.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 1.0 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 5.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 8.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 5.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 10.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 9.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 2.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 7.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 3.6 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 8.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 4.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 3.8 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 4.3 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 5.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 2.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 2.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 4.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.4 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.4 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 24.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.5 | 10.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.4 | 8.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.3 | 1.4 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.2 | 5.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 5.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 4.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.2 | 2.7 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 2.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 3.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 2.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 4.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 3.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 3.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 3.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 4.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 8.8 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 1.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 2.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 2.0 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 3.1 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 2.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.9 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.1 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.9 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 5.3 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.8 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 1.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.7 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 1.4 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 3.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR1 MUTANTS | Genes involved in Signaling by FGFR1 mutants |
| 0.0 | 0.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.3 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 1.6 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 4.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 3.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 1.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.8 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |