Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Sox7
Z-value: 0.68
Transcription factors associated with Sox7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox7
|
ENSMUSG00000063060.7 | Sox7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox7 | mm39_v1_chr14_+_64181115_64181136 | -0.15 | 2.0e-01 | Click! |
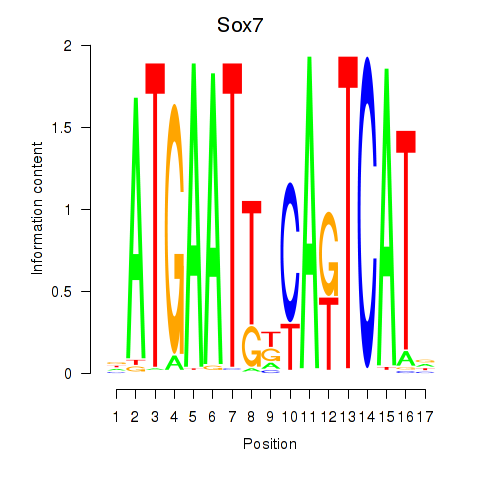
Activity profile of Sox7 motif
Sorted Z-values of Sox7 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox7
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_92840279 | 5.85 |
ENSMUST00000046234.5
|
Lce3b
|
late cornified envelope 3B |
| chr7_-_14180496 | 4.33 |
ENSMUST00000063509.11
|
Sult2a8
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
| chr7_-_14180576 | 4.29 |
ENSMUST00000125941.3
|
Sult2a8
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
| chrX_+_102465616 | 3.65 |
ENSMUST00000182089.2
|
Gm26992
|
predicted gene, 26992 |
| chr3_+_146302832 | 3.55 |
ENSMUST00000029837.14
ENSMUST00000147409.2 ENSMUST00000121133.2 |
Uox
|
urate oxidase |
| chr3_-_69767208 | 3.54 |
ENSMUST00000171529.4
ENSMUST00000051239.13 |
Sptssb
|
serine palmitoyltransferase, small subunit B |
| chr1_+_107288928 | 3.48 |
ENSMUST00000191425.7
|
Serpinb11
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 11 |
| chr8_-_62576140 | 3.42 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr1_+_107289659 | 2.97 |
ENSMUST00000027566.3
|
Serpinb11
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 11 |
| chr1_-_119576347 | 2.93 |
ENSMUST00000027632.14
ENSMUST00000187194.2 |
Epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr6_+_48661503 | 2.89 |
ENSMUST00000067506.14
ENSMUST00000119575.8 ENSMUST00000121957.8 ENSMUST00000090070.6 |
Gimap4
|
GTPase, IMAP family member 4 |
| chr2_-_6134926 | 2.89 |
ENSMUST00000126551.2
ENSMUST00000054254.12 ENSMUST00000114942.9 |
Proser2
|
proline and serine rich 2 |
| chr2_-_120144622 | 2.63 |
ENSMUST00000054651.8
|
Pla2g4f
|
phospholipase A2, group IVF |
| chr3_+_59914164 | 2.58 |
ENSMUST00000169794.2
|
Aadacl2
|
arylacetamide deacetylase like 2 |
| chr11_-_46280336 | 2.55 |
ENSMUST00000020664.13
|
Itk
|
IL2 inducible T cell kinase |
| chr6_+_48661477 | 2.47 |
ENSMUST00000118802.8
|
Gimap4
|
GTPase, IMAP family member 4 |
| chr12_-_113379925 | 2.45 |
ENSMUST00000194162.6
ENSMUST00000192250.2 |
Ighd
|
immunoglobulin heavy constant delta |
| chr11_-_46280298 | 2.42 |
ENSMUST00000109237.9
|
Itk
|
IL2 inducible T cell kinase |
| chr2_-_93283024 | 2.28 |
ENSMUST00000111257.8
ENSMUST00000145553.8 |
Cd82
|
CD82 antigen |
| chr12_-_75782406 | 2.28 |
ENSMUST00000220285.2
|
Sgpp1
|
sphingosine-1-phosphate phosphatase 1 |
| chr12_-_75782502 | 2.23 |
ENSMUST00000021450.6
|
Sgpp1
|
sphingosine-1-phosphate phosphatase 1 |
| chr11_-_46057224 | 2.10 |
ENSMUST00000020679.3
|
Nipal4
|
NIPA-like domain containing 4 |
| chr6_-_68713748 | 1.85 |
ENSMUST00000183936.2
ENSMUST00000196863.2 |
Igkv19-93
|
immunoglobulin kappa chain variable 19-93 |
| chr3_-_146302343 | 1.84 |
ENSMUST00000029836.9
|
Dnase2b
|
deoxyribonuclease II beta |
| chr16_+_8455538 | 1.62 |
ENSMUST00000023396.10
ENSMUST00000230828.2 |
Pmm2
|
phosphomannomutase 2 |
| chr11_-_46280281 | 1.62 |
ENSMUST00000101306.4
|
Itk
|
IL2 inducible T cell kinase |
| chr6_+_67532481 | 1.51 |
ENSMUST00000103302.3
|
Igkv2-137
|
immunoglobulin kappa chain variable 2-137 |
| chr3_+_90561560 | 1.18 |
ENSMUST00000079286.4
|
S100a7a
|
S100 calcium binding protein A7A |
| chr6_-_99243455 | 1.17 |
ENSMUST00000113326.9
|
Foxp1
|
forkhead box P1 |
| chr8_+_123829089 | 1.11 |
ENSMUST00000000756.6
|
Rpl13
|
ribosomal protein L13 |
| chr1_-_173363523 | 1.08 |
ENSMUST00000139092.8
|
Ifi214
|
interferon activated gene 214 |
| chr3_-_30194559 | 0.92 |
ENSMUST00000108271.10
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr7_-_6502647 | 0.88 |
ENSMUST00000209055.2
ENSMUST00000207339.4 |
Olfr1347
|
olfactory receptor 1347 |
| chr1_-_171476559 | 0.83 |
ENSMUST00000111276.9
ENSMUST00000194531.6 |
Slamf7
|
SLAM family member 7 |
| chr13_-_4200627 | 0.81 |
ENSMUST00000110704.9
ENSMUST00000021635.9 |
Akr1c18
|
aldo-keto reductase family 1, member C18 |
| chr1_-_171476495 | 0.79 |
ENSMUST00000194791.2
ENSMUST00000192024.6 |
Slamf7
|
SLAM family member 7 |
| chr14_+_54433524 | 0.75 |
ENSMUST00000103718.2
|
Traj23
|
T cell receptor alpha joining 23 |
| chr9_-_111100859 | 0.74 |
ENSMUST00000035079.10
|
Mlh1
|
mutL homolog 1 |
| chr14_+_54433095 | 0.74 |
ENSMUST00000103717.3
|
Traj24
|
T cell receptor alpha joining 24 |
| chr2_+_15060051 | 0.72 |
ENSMUST00000069870.11
ENSMUST00000239125.2 ENSMUST00000193836.3 |
Arl5b
|
ADP-ribosylation factor-like 5B |
| chr19_-_32085532 | 0.70 |
ENSMUST00000236030.2
ENSMUST00000237104.2 ENSMUST00000236208.2 ENSMUST00000235273.2 |
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr2_-_86640362 | 0.64 |
ENSMUST00000216117.2
|
Olfr141
|
olfactory receptor 141 |
| chr10_-_107747995 | 0.56 |
ENSMUST00000165341.5
|
Otogl
|
otogelin-like |
| chr9_-_36678868 | 0.53 |
ENSMUST00000217599.2
ENSMUST00000120381.9 |
Stt3a
|
STT3, subunit of the oligosaccharyltransferase complex, homolog A (S. cerevisiae) |
| chr14_+_64890008 | 0.48 |
ENSMUST00000224503.2
|
Kif13b
|
kinesin family member 13B |
| chr19_-_12881094 | 0.45 |
ENSMUST00000208343.2
ENSMUST00000216989.2 |
Olfr1447
|
olfactory receptor 1447 |
| chr2_+_38401655 | 0.44 |
ENSMUST00000054234.10
ENSMUST00000112902.8 |
Nek6
|
NIMA (never in mitosis gene a)-related expressed kinase 6 |
| chr1_-_136888118 | 0.43 |
ENSMUST00000192357.6
ENSMUST00000027649.14 |
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr7_-_10488291 | 0.42 |
ENSMUST00000226874.2
ENSMUST00000227003.2 ENSMUST00000228561.2 ENSMUST00000228248.2 ENSMUST00000228526.2 ENSMUST00000228098.2 ENSMUST00000227940.2 ENSMUST00000228374.2 ENSMUST00000227702.2 |
Vmn1r71
|
vomeronasal 1 receptor 71 |
| chr9_-_35559489 | 0.37 |
ENSMUST00000178236.3
|
Pate3
|
prostate and testis expressed 3 |
| chr5_-_108022900 | 0.34 |
ENSMUST00000138111.8
ENSMUST00000112642.8 |
Evi5
|
ecotropic viral integration site 5 |
| chr17_+_38143840 | 0.28 |
ENSMUST00000213857.2
|
Olfr125
|
olfactory receptor 125 |
| chr2_+_88660081 | 0.24 |
ENSMUST00000215929.2
|
Olfr1205
|
olfactory receptor 1205 |
| chr7_-_7822866 | 0.24 |
ENSMUST00000169683.2
|
Vmn2r35
|
vomeronasal 2, receptor 35 |
| chr3_-_151468529 | 0.21 |
ENSMUST00000046739.6
|
Ifi44l
|
interferon-induced protein 44 like |
| chr12_-_114793177 | 0.19 |
ENSMUST00000103511.2
ENSMUST00000195735.2 |
Ighv1-31
|
immunoglobulin heavy variable 1-31 |
| chr7_-_7569785 | 0.17 |
ENSMUST00000165921.2
|
Vmn2r33
|
vomeronasal 2, receptor 33 |
| chr19_+_13717092 | 0.13 |
ENSMUST00000207836.3
|
Olfr1494
|
olfactory receptor 1494 |
| chr11_+_73489420 | 0.11 |
ENSMUST00000214228.2
|
Olfr384
|
olfactory receptor 384 |
| chr9_+_19495807 | 0.10 |
ENSMUST00000061693.5
|
Olfr855
|
olfactory receptor 855 |
| chr7_-_119078472 | 0.09 |
ENSMUST00000209095.2
ENSMUST00000033263.6 ENSMUST00000207261.2 |
Umod
|
uromodulin |
| chr14_+_64889973 | 0.09 |
ENSMUST00000100473.5
|
Kif13b
|
kinesin family member 13B |
| chr16_-_58583539 | 0.09 |
ENSMUST00000214139.2
|
Olfr172
|
olfactory receptor 172 |
| chr14_+_66378382 | 0.06 |
ENSMUST00000022620.11
|
Chrna2
|
cholinergic receptor, nicotinic, alpha polypeptide 2 (neuronal) |
| chr7_-_106279791 | 0.05 |
ENSMUST00000215541.2
|
Olfr693
|
olfactory receptor 693 |
| chr7_+_45164042 | 0.05 |
ENSMUST00000210868.2
|
Tulp2
|
tubby-like protein 2 |
| chr2_-_111104451 | 0.03 |
ENSMUST00000214760.2
|
Olfr1277
|
olfactory receptor 1277 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.8 | 4.5 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.4 | 3.5 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.4 | 1.6 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.3 | 6.6 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.3 | 0.8 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.2 | 3.2 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.2 | 0.5 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.2 | 3.6 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 1.2 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 2.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 2.6 | GO:0071236 | cellular response to antibiotic(GO:0071236) |
| 0.1 | 5.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 1.8 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 0.9 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.6 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.4 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 1.6 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) |
| 0.0 | 5.9 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 3.0 | GO:0002377 | immunoglobulin production(GO:0002377) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.5 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.2 | 0.7 | GO:0005712 | chiasma(GO:0005712) late recombination nodule(GO:0005715) |
| 0.2 | 2.5 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 5.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 5.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 6.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 3.6 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.6 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 1.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.5 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.7 | 3.5 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.5 | 1.6 | GO:0004615 | phosphomannomutase activity(GO:0004615) |
| 0.4 | 1.8 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.3 | 2.6 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.3 | 3.6 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.3 | 0.8 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.2 | 0.7 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.1 | 2.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.7 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 6.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.5 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 6.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 1.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 2.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 2.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 6.1 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 6.6 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 2.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 4.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | PID ARF6 PATHWAY | Arf6 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 2.6 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 4.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |