Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
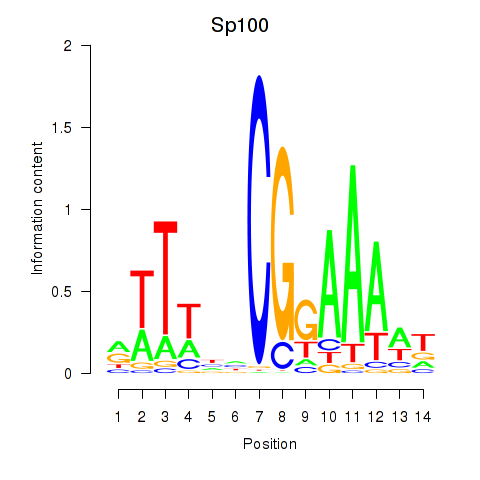
Results for Sp100
Z-value: 0.85
Transcription factors associated with Sp100
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sp100
|
ENSMUSG00000026222.17 | Sp100 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sp100 | mm39_v1_chr1_+_85577766_85577774 | -0.13 | 2.6e-01 | Click! |
Activity profile of Sp100 motif
Sorted Z-values of Sp100 motif
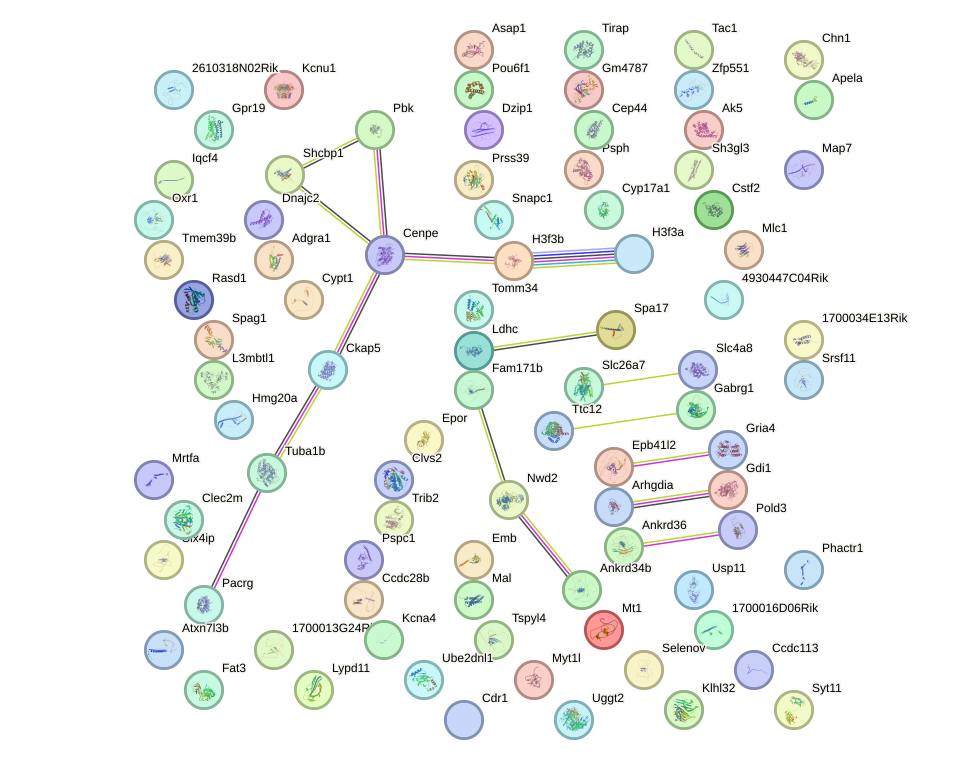
Network of associatons between targets according to the STRING database.
First level regulatory network of Sp100
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_60229164 | 6.12 |
ENSMUST00000166381.3
|
Cdr1
|
cerebellar degeneration related antigen 1 |
| chr9_-_4796217 | 6.11 |
ENSMUST00000027020.13
ENSMUST00000163309.2 |
Gria4
|
glutamate receptor, ionotropic, AMPA4 (alpha 4) |
| chr3_-_88679881 | 4.73 |
ENSMUST00000090945.5
|
Syt11
|
synaptotagmin XI |
| chrX_+_20570145 | 4.64 |
ENSMUST00000033383.3
|
Usp11
|
ubiquitin specific peptidase 11 |
| chr7_-_24423715 | 4.23 |
ENSMUST00000081657.6
|
Lypd11
|
Ly6/PLAUR domain containing 11 |
| chr3_-_33039225 | 4.16 |
ENSMUST00000108221.2
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr6_-_129308748 | 4.12 |
ENSMUST00000051283.8
|
Clec2m
|
C-type lectin domain family 2, member m |
| chr16_-_16942970 | 4.02 |
ENSMUST00000093336.8
ENSMUST00000231681.2 |
2610318N02Rik
|
RIKEN cDNA 2610318N02 gene |
| chr15_-_88863210 | 3.96 |
ENSMUST00000042594.13
ENSMUST00000109368.2 |
Mlc1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 homolog (human) |
| chr14_+_66043515 | 3.83 |
ENSMUST00000139644.2
|
Pbk
|
PDZ binding kinase |
| chr2_+_83642910 | 3.67 |
ENSMUST00000051454.4
|
Fam171b
|
family with sequence similarity 171, member B |
| chr13_+_43019718 | 3.45 |
ENSMUST00000131942.2
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr7_+_81824544 | 3.39 |
ENSMUST00000032874.14
|
Sh3gl3
|
SH3-domain GRB2-like 3 |
| chr7_+_139414057 | 3.37 |
ENSMUST00000026548.14
|
Adgra1
|
adhesion G protein-coupled receptor A1 |
| chr8_+_94905710 | 3.26 |
ENSMUST00000034215.8
ENSMUST00000212291.2 ENSMUST00000211807.2 |
Mt1
|
metallothionein 1 |
| chr7_+_46510627 | 3.07 |
ENSMUST00000014545.11
|
Ldhc
|
lactate dehydrogenase C |
| chr10_-_33500583 | 2.87 |
ENSMUST00000161692.2
ENSMUST00000160299.2 ENSMUST00000019920.13 |
Clvs2
|
clavesin 2 |
| chr3_-_152232389 | 2.77 |
ENSMUST00000200062.2
|
Ak5
|
adenylate kinase 5 |
| chr14_-_119336809 | 2.66 |
ENSMUST00000156203.8
|
Uggt2
|
UDP-glucose glycoprotein glucosyltransferase 2 |
| chr8_+_96260694 | 2.61 |
ENSMUST00000041569.5
|
Ccdc113
|
coiled-coil domain containing 113 |
| chr13_+_92562404 | 2.54 |
ENSMUST00000061594.13
|
Ankrd34b
|
ankyrin repeat domain 34B |
| chr10_+_34173426 | 2.52 |
ENSMUST00000047935.8
|
Tspyl4
|
TSPY-like 4 |
| chr2_-_127498129 | 2.51 |
ENSMUST00000028853.7
|
Mal
|
myelin and lymphocyte protein, T cell differentiation protein |
| chr2_-_73605387 | 2.48 |
ENSMUST00000166199.9
|
Chn1
|
chimerin 1 |
| chr5_-_70999547 | 2.48 |
ENSMUST00000199705.2
|
Gabrg1
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 1 |
| chr3_+_134918298 | 2.47 |
ENSMUST00000062893.12
|
Cenpe
|
centromere protein E |
| chr4_+_137180565 | 2.46 |
ENSMUST00000048893.4
|
1700013G24Rik
|
RIKEN cDNA 1700013G24 gene |
| chr13_-_3661064 | 2.45 |
ENSMUST00000096069.5
|
Tasor2
|
transcription activation suppressor family member 2 |
| chr14_-_119160497 | 2.34 |
ENSMUST00000047208.12
|
Dzip1
|
DAZ interacting protein 1 |
| chr11_-_69706124 | 2.34 |
ENSMUST00000210714.2
|
Gm39566
|
predicted gene, 39566 |
| chr12_+_29584560 | 2.33 |
ENSMUST00000021009.10
|
Myt1l
|
myelin transcription factor 1-like |
| chr7_+_127172701 | 2.31 |
ENSMUST00000121004.3
|
Phkg2
|
phosphorylase kinase, gamma 2 (testis) |
| chr17_-_11059172 | 2.26 |
ENSMUST00000041463.7
|
Pacrg
|
PARK2 co-regulated |
| chr13_+_49806772 | 2.24 |
ENSMUST00000223264.2
ENSMUST00000221142.2 ENSMUST00000222333.2 ENSMUST00000021824.8 |
Nol8
|
nucleolar protein 8 |
| chr8_+_26339646 | 2.24 |
ENSMUST00000098858.11
|
Kcnu1
|
potassium channel, subfamily U, member 1 |
| chr18_+_37125424 | 2.23 |
ENSMUST00000194038.2
|
Pcdha8
|
protocadherin alpha 8 |
| chr18_+_52779281 | 2.23 |
ENSMUST00000118724.8
ENSMUST00000091904.6 |
1700034E13Rik
|
RIKEN cDNA 1700034E13 gene |
| chr15_+_100659622 | 2.20 |
ENSMUST00000023776.13
|
Slc4a8
|
solute carrier family 4 (anion exchanger), member 8 |
| chr2_+_91376650 | 2.20 |
ENSMUST00000099716.11
ENSMUST00000046769.16 ENSMUST00000111337.3 |
Ckap5
|
cytoskeleton associated protein 5 |
| chrX_+_152341587 | 2.17 |
ENSMUST00000112573.8
ENSMUST00000056754.4 |
Cypt3
|
cysteine-rich perinuclear theca 3 |
| chr13_+_117356808 | 2.17 |
ENSMUST00000022242.9
|
Emb
|
embigin |
| chr2_-_163913014 | 2.15 |
ENSMUST00000018466.4
ENSMUST00000109384.10 |
Tomm34
|
translocase of outer mitochondrial membrane 34 |
| chr7_-_27990555 | 2.13 |
ENSMUST00000056589.15
|
Selenov
|
selenoprotein V |
| chr17_-_10501816 | 2.10 |
ENSMUST00000233684.2
|
Qk
|
quaking, KH domain containing RNA binding |
| chr12_-_81426238 | 2.08 |
ENSMUST00000062182.8
|
Gm4787
|
predicted gene 4787 |
| chrX_+_113814692 | 2.08 |
ENSMUST00000059509.3
|
Ube2dnl1
|
ubiquitin-conjugating enzyme E2D N-terminal like 1 |
| chr17_+_47816074 | 2.07 |
ENSMUST00000183177.8
ENSMUST00000182848.8 |
Ccnd3
|
cyclin D3 |
| chr2_-_111059901 | 2.05 |
ENSMUST00000028577.3
|
A26c3
|
ANKRD26-like family C, member 3 |
| chrX_+_16389108 | 2.04 |
ENSMUST00000115422.2
ENSMUST00000024026.3 |
Cypt1
|
cysteine-rich perinuclear theca 1 |
| chr8_-_4829519 | 2.03 |
ENSMUST00000022945.9
|
Shcbp1
|
Shc SH2-domain binding protein 1 |
| chr8_-_11728727 | 2.03 |
ENSMUST00000033906.11
|
1700016D06Rik
|
RIKEN cDNA 1700016D06 gene |
| chr4_-_119217079 | 2.00 |
ENSMUST00000143494.3
ENSMUST00000154606.9 |
Ccdc30
|
coiled-coil domain containing 30 |
| chr9_-_106448182 | 2.00 |
ENSMUST00000085111.5
|
Iqcf4
|
IQ motif containing F4 |
| chr12_-_72964646 | 1.99 |
ENSMUST00000044000.12
|
4930447C04Rik
|
RIKEN cDNA 4930447C04 gene |
| chr9_-_49397508 | 1.97 |
ENSMUST00000055096.5
|
Ttc12
|
tetratricopeptide repeat domain 12 |
| chr9_-_4795519 | 1.97 |
ENSMUST00000212533.2
|
Gria4
|
glutamate receptor, ionotropic, AMPA4 (alpha 4) |
| chr5_-_21990170 | 1.96 |
ENSMUST00000115193.8
ENSMUST00000115192.2 ENSMUST00000115195.8 ENSMUST00000030771.12 |
Dnajc2
|
DnaJ heat shock protein family (Hsp40) member C2 |
| chr16_+_7011580 | 1.91 |
ENSMUST00000231194.2
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr18_-_23171713 | 1.90 |
ENSMUST00000081423.13
|
Nol4
|
nucleolar protein 4 |
| chr5_+_63806451 | 1.89 |
ENSMUST00000159584.3
|
Nwd2
|
NACHT and WD repeat domain containing 2 |
| chr15_-_98832403 | 1.88 |
ENSMUST00000077577.8
|
Tuba1b
|
tubulin, alpha 1B |
| chr6_-_134874778 | 1.88 |
ENSMUST00000165392.8
ENSMUST00000204880.3 ENSMUST00000203409.3 ENSMUST00000046255.14 ENSMUST00000111932.8 ENSMUST00000149375.8 ENSMUST00000116515.9 |
Gpr19
|
G protein-coupled receptor 19 |
| chr19_-_46661501 | 1.86 |
ENSMUST00000236174.2
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr2_+_136734088 | 1.84 |
ENSMUST00000099311.9
|
Slx4ip
|
SLX4 interacting protein |
| chr9_-_21874802 | 1.83 |
ENSMUST00000006397.7
|
Epor
|
erythropoietin receptor |
| chr15_+_41573995 | 1.81 |
ENSMUST00000229769.2
|
Oxr1
|
oxidation resistance 1 |
| chr18_+_37884653 | 1.81 |
ENSMUST00000194928.2
|
Pcdhgb7
|
protocadherin gamma subfamily B, 7 |
| chr10_+_25284244 | 1.79 |
ENSMUST00000092645.7
ENSMUST00000219805.2 |
Epb41l2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr2_+_162785394 | 1.78 |
ENSMUST00000035751.12
ENSMUST00000156954.8 |
L3mbtl1
|
L3MBTL1 histone methyl-lysine binding protein |
| chr11_-_59855762 | 1.78 |
ENSMUST00000062405.8
|
Rasd1
|
RAS, dexamethasone-induced 1 |
| chr9_-_37525009 | 1.77 |
ENSMUST00000002013.11
|
Spa17
|
sperm autoantigenic protein 17 |
| chr4_-_129517662 | 1.75 |
ENSMUST00000106035.8
ENSMUST00000150357.2 ENSMUST00000030586.15 |
Ccdc28b
|
coiled coil domain containing 28B |
| chr4_-_129334593 | 1.75 |
ENSMUST00000053042.6
ENSMUST00000106046.8 |
Zbtb8b
|
zinc finger and BTB domain containing 8b |
| chr15_-_64254754 | 1.75 |
ENSMUST00000177374.8
ENSMUST00000110114.10 ENSMUST00000110115.9 ENSMUST00000023008.16 |
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
| chr10_-_103072141 | 1.74 |
ENSMUST00000123364.2
ENSMUST00000166240.8 ENSMUST00000020043.12 |
Lrriq1
|
leucine-rich repeats and IQ motif containing 1 |
| chr1_+_34537450 | 1.74 |
ENSMUST00000027299.10
|
Prss39
|
protease, serine 39 |
| chr2_-_18806629 | 1.74 |
ENSMUST00000181515.2
|
Carlr
|
cardiac and apoptosis-related long non-coding RNA |
| chr3_-_157742204 | 1.73 |
ENSMUST00000137444.8
|
Srsf11
|
serine and arginine-rich splicing factor 11 |
| chr8_-_57003828 | 1.69 |
ENSMUST00000134162.8
ENSMUST00000140107.8 ENSMUST00000040330.15 ENSMUST00000135337.8 |
Cep44
|
centrosomal protein 44 |
| chr9_+_56325893 | 1.68 |
ENSMUST00000034879.5
ENSMUST00000215269.2 |
Hmg20a
|
high mobility group 20A |
| chr3_-_157742182 | 1.67 |
ENSMUST00000069025.14
|
Srsf11
|
serine and arginine-rich splicing factor 11 |
| chr15_-_64254585 | 1.66 |
ENSMUST00000176384.8
ENSMUST00000175799.8 |
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
| chr4_-_24851086 | 1.66 |
ENSMUST00000084781.6
ENSMUST00000108218.10 |
Klhl32
|
kelch-like 32 |
| chr15_+_36179676 | 1.65 |
ENSMUST00000171205.3
|
Spag1
|
sperm associated antigen 1 |
| chr10_-_112764879 | 1.65 |
ENSMUST00000099276.4
|
Atxn7l3b
|
ataxin 7-like 3B |
| chrX_+_73352694 | 1.65 |
ENSMUST00000130581.2
|
Gdi1
|
guanosine diphosphate (GDP) dissociation inhibitor 1 |
| chr3_-_157742267 | 1.64 |
ENSMUST00000121326.8
|
Srsf11
|
serine and arginine-rich splicing factor 11 |
| chr6_+_7555053 | 1.62 |
ENSMUST00000090679.9
ENSMUST00000184986.2 |
Tac1
|
tachykinin 1 |
| chr7_-_12156516 | 1.62 |
ENSMUST00000120220.3
|
Zfp551
|
zinc fingr protein 551 |
| chr12_+_74011255 | 1.62 |
ENSMUST00000021532.6
|
Snapc1
|
small nuclear RNA activating complex, polypeptide 1 |
| chr4_-_129590609 | 1.60 |
ENSMUST00000102588.10
|
Tmem39b
|
transmembrane protein 39b |
| chr5_-_129864202 | 1.59 |
ENSMUST00000136507.4
|
Psph
|
phosphoserine phosphatase |
| chrX_+_132959905 | 1.56 |
ENSMUST00000113287.8
ENSMUST00000033609.9 ENSMUST00000113286.8 |
Cstf2
|
cleavage stimulation factor, 3' pre-RNA subunit 2 |
| chr8_-_65489834 | 1.52 |
ENSMUST00000142822.4
|
Apela
|
apelin receptor early endogenous ligand |
| chr7_+_5059703 | 1.52 |
ENSMUST00000208042.2
ENSMUST00000207974.2 |
Ccdc106
|
coiled-coil domain containing 106 |
| chr13_+_49806542 | 1.52 |
ENSMUST00000222197.2
ENSMUST00000221083.2 ENSMUST00000223467.2 |
Nol8
|
nucleolar protein 8 |
| chr2_+_107120934 | 1.51 |
ENSMUST00000037012.3
|
Kcna4
|
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr11_+_5519677 | 1.50 |
ENSMUST00000109856.8
ENSMUST00000109855.8 ENSMUST00000118112.9 |
Ankrd36
|
ankyrin repeat domain 36 |
| chr4_-_14621805 | 1.49 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr10_+_20024203 | 1.49 |
ENSMUST00000020173.16
|
Map7
|
microtubule-associated protein 7 |
| chr11_-_115915315 | 1.47 |
ENSMUST00000016703.8
|
H3f3b
|
H3.3 histone B |
| chr14_-_57015748 | 1.46 |
ENSMUST00000022507.13
ENSMUST00000163924.2 |
Pspc1
|
paraspeckle protein 1 |
| chr9_-_16289527 | 1.45 |
ENSMUST00000082170.6
|
Fat3
|
FAT atypical cadherin 3 |
| chr7_-_99770653 | 1.45 |
ENSMUST00000208670.2
ENSMUST00000032969.14 |
Pold3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr12_-_15866763 | 1.44 |
ENSMUST00000020922.8
ENSMUST00000221215.2 ENSMUST00000221518.2 |
Trib2
|
tribbles pseudokinase 2 |
| chr2_+_71042285 | 1.44 |
ENSMUST00000112144.9
ENSMUST00000100028.10 ENSMUST00000112136.2 |
Dync1i2
|
dynein cytoplasmic 1 intermediate chain 2 |
| chr15_-_74508197 | 1.43 |
ENSMUST00000023271.8
|
Mroh4
|
maestro heat-like repeat family member 4 |
| chrX_+_52726793 | 1.42 |
ENSMUST00000069209.2
|
Ct55
|
cancer/testis antigen 55 |
| chr13_+_112797273 | 1.38 |
ENSMUST00000052514.6
|
Slc38a9
|
solute carrier family 38, member 9 |
| chr17_+_47816137 | 1.35 |
ENSMUST00000182935.8
ENSMUST00000182506.8 |
Ccnd3
|
cyclin D3 |
| chr6_+_96090127 | 1.33 |
ENSMUST00000122120.8
|
Tafa1
|
TAFA chemokine like family member 1 |
| chr2_+_136374186 | 1.32 |
ENSMUST00000121717.2
|
Ankef1
|
ankyrin repeat and EF-hand domain containing 1 |
| chr5_+_150597292 | 1.30 |
ENSMUST00000038900.15
|
Pds5b
|
PDS5 cohesin associated factor B |
| chr4_-_129512862 | 1.30 |
ENSMUST00000121442.2
ENSMUST00000046675.12 |
Iqcc
|
IQ motif containing C |
| chr12_+_98737274 | 1.28 |
ENSMUST00000021399.9
|
Zc3h14
|
zinc finger CCCH type containing 14 |
| chr1_+_82702598 | 1.28 |
ENSMUST00000078332.13
ENSMUST00000073025.12 ENSMUST00000161648.8 ENSMUST00000160786.8 ENSMUST00000162003.8 |
Mff
|
mitochondrial fission factor |
| chr8_+_70735477 | 1.27 |
ENSMUST00000087467.12
ENSMUST00000140212.8 ENSMUST00000110124.9 |
Homer3
|
homer scaffolding protein 3 |
| chr18_+_63110899 | 1.26 |
ENSMUST00000025474.14
|
Napg
|
N-ethylmaleimide sensitive fusion protein attachment protein gamma |
| chr13_-_49806231 | 1.26 |
ENSMUST00000021818.9
|
Cenpp
|
centromere protein P |
| chr18_+_63111005 | 1.25 |
ENSMUST00000235372.2
ENSMUST00000237483.2 |
Napg
|
N-ethylmaleimide sensitive fusion protein attachment protein gamma |
| chr15_+_34837501 | 1.24 |
ENSMUST00000072868.5
|
Kcns2
|
K+ voltage-gated channel, subfamily S, 2 |
| chr8_-_84791766 | 1.23 |
ENSMUST00000098592.3
|
Gm10643
|
predicted gene 10643 |
| chr7_-_12096691 | 1.23 |
ENSMUST00000086228.3
|
Vmn1r84
|
vomeronasal 1 receptor 84 |
| chr12_-_110649040 | 1.21 |
ENSMUST00000222915.2
ENSMUST00000070659.7 |
1700001K19Rik
|
RIKEN cDNA 1700001K19 gene |
| chr12_-_46767619 | 1.20 |
ENSMUST00000219886.2
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr11_+_109317046 | 1.20 |
ENSMUST00000238825.2
ENSMUST00000103061.3 |
Amz2
|
archaelysin family metallopeptidase 2 |
| chr13_+_18901459 | 1.18 |
ENSMUST00000072961.6
|
Vps41
|
VPS41 HOPS complex subunit |
| chr11_-_78642480 | 1.17 |
ENSMUST00000059468.6
|
Ccnq
|
cyclin Q |
| chr18_+_63110924 | 1.17 |
ENSMUST00000150267.2
ENSMUST00000236925.2 |
Napg
|
N-ethylmaleimide sensitive fusion protein attachment protein gamma |
| chr7_-_43906802 | 1.15 |
ENSMUST00000107945.8
ENSMUST00000118216.8 |
Acp4
|
acid phosphatase 4 |
| chr18_-_12996348 | 1.15 |
ENSMUST00000143077.8
ENSMUST00000117361.8 ENSMUST00000234871.2 |
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr8_-_87307294 | 1.14 |
ENSMUST00000131423.8
ENSMUST00000152438.2 |
Abcc12
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 12 |
| chr9_+_26941945 | 1.12 |
ENSMUST00000216677.2
|
Ncapd3
|
non-SMC condensin II complex, subunit D3 |
| chr7_-_126101245 | 1.11 |
ENSMUST00000179818.3
|
Atxn2l
|
ataxin 2-like |
| chr2_-_17465410 | 1.10 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr5_+_123648523 | 1.10 |
ENSMUST00000031384.6
|
B3gnt4
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr15_+_100178718 | 1.10 |
ENSMUST00000096200.6
|
Tmprss12
|
transmembrane (C-terminal) protease, serine 12 |
| chr2_+_71042172 | 1.10 |
ENSMUST00000081710.12
|
Dync1i2
|
dynein cytoplasmic 1 intermediate chain 2 |
| chr2_-_91067212 | 1.10 |
ENSMUST00000111352.8
|
Ddb2
|
damage specific DNA binding protein 2 |
| chr1_-_60137294 | 1.10 |
ENSMUST00000141417.3
ENSMUST00000122038.8 |
Wdr12
|
WD repeat domain 12 |
| chr8_-_26087552 | 1.09 |
ENSMUST00000210234.2
ENSMUST00000211422.2 |
Letm2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr2_+_71042050 | 1.08 |
ENSMUST00000112142.8
ENSMUST00000112139.8 ENSMUST00000112140.8 ENSMUST00000112138.8 |
Dync1i2
|
dynein cytoplasmic 1 intermediate chain 2 |
| chr3_+_40978804 | 1.08 |
ENSMUST00000099121.10
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr12_+_85871691 | 1.06 |
ENSMUST00000040273.14
ENSMUST00000110224.8 ENSMUST00000142411.9 |
Ttll5
|
tubulin tyrosine ligase-like family, member 5 |
| chr8_+_31640332 | 1.04 |
ENSMUST00000209851.2
ENSMUST00000098842.3 ENSMUST00000210129.2 |
Tti2
|
TELO2 interacting protein 2 |
| chr13_+_104953679 | 1.04 |
ENSMUST00000022230.15
|
Srek1ip1
|
splicing regulatory glutamine/lysine-rich protein 1interacting protein 1 |
| chr5_+_150597204 | 1.04 |
ENSMUST00000202170.4
ENSMUST00000016569.11 |
Pds5b
|
PDS5 cohesin associated factor B |
| chr16_-_4536943 | 1.02 |
ENSMUST00000120056.8
ENSMUST00000074970.8 |
Nmral1
|
NmrA-like family domain containing 1 |
| chr18_+_37864045 | 1.02 |
ENSMUST00000192535.2
|
Pcdhgb5
|
protocadherin gamma subfamily B, 5 |
| chr9_+_3335135 | 0.98 |
ENSMUST00000212294.2
|
Alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr2_+_102489558 | 0.98 |
ENSMUST00000111213.8
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chrX_-_65347749 | 0.96 |
ENSMUST00000033525.3
|
4930447F04Rik
|
RIKEN cDNA 4930447F04 gene |
| chr8_-_65489791 | 0.95 |
ENSMUST00000124790.8
|
Apela
|
apelin receptor early endogenous ligand |
| chr16_+_17328924 | 0.95 |
ENSMUST00000232372.2
|
Lztr1
|
leucine-zipper-like transcriptional regulator, 1 |
| chr11_+_103857541 | 0.94 |
ENSMUST00000057921.10
ENSMUST00000063347.12 |
Arf2
|
ADP-ribosylation factor 2 |
| chr4_+_33310306 | 0.91 |
ENSMUST00000108153.9
ENSMUST00000029942.8 |
Rngtt
|
RNA guanylyltransferase and 5'-phosphatase |
| chr17_-_36149100 | 0.88 |
ENSMUST00000134978.3
|
Tubb5
|
tubulin, beta 5 class I |
| chr6_-_106777014 | 0.88 |
ENSMUST00000013882.10
ENSMUST00000113239.10 |
Crbn
|
cereblon |
| chr7_-_79570342 | 0.87 |
ENSMUST00000075657.8
|
Ap3s2
|
adaptor-related protein complex 3, sigma 2 subunit |
| chr2_-_151583155 | 0.86 |
ENSMUST00000042452.11
|
Psmf1
|
proteasome (prosome, macropain) inhibitor subunit 1 |
| chr2_-_112198366 | 0.86 |
ENSMUST00000028551.4
|
Emc4
|
ER membrane protein complex subunit 4 |
| chr13_-_23946359 | 0.85 |
ENSMUST00000091701.3
|
H3c1
|
H3 clustered histone 1 |
| chr18_+_37580692 | 0.85 |
ENSMUST00000052387.5
|
Pcdhb14
|
protocadherin beta 14 |
| chr1_-_118239146 | 0.85 |
ENSMUST00000027623.9
|
Tsn
|
translin |
| chr17_-_47922374 | 0.84 |
ENSMUST00000024783.9
|
Bysl
|
bystin-like |
| chr5_+_21990251 | 0.82 |
ENSMUST00000239497.2
ENSMUST00000030769.7 |
Psmc2
|
proteasome (prosome, macropain) 26S subunit, ATPase 2 |
| chr13_+_21364330 | 0.81 |
ENSMUST00000223065.2
|
Trim27
|
tripartite motif-containing 27 |
| chr9_-_36637670 | 0.81 |
ENSMUST00000172702.9
ENSMUST00000172742.2 |
Chek1
|
checkpoint kinase 1 |
| chrX_+_67722147 | 0.80 |
ENSMUST00000088546.12
|
Fmr1
|
FMRP translational regulator 1 |
| chr9_+_56326089 | 0.80 |
ENSMUST00000213242.2
ENSMUST00000214771.2 ENSMUST00000217518.2 ENSMUST00000214869.2 |
Hmg20a
|
high mobility group 20A |
| chr5_+_25451771 | 0.79 |
ENSMUST00000144971.2
|
Galnt11
|
polypeptide N-acetylgalactosaminyltransferase 11 |
| chr11_-_70242850 | 0.79 |
ENSMUST00000019068.7
|
Alox15
|
arachidonate 15-lipoxygenase |
| chr7_-_13856967 | 0.79 |
ENSMUST00000098809.4
|
Sult2a3
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 3 |
| chr19_+_4806544 | 0.78 |
ENSMUST00000182821.8
ENSMUST00000036744.8 |
Rbm4b
|
RNA binding motif protein 4B |
| chr9_+_3335469 | 0.78 |
ENSMUST00000053407.13
ENSMUST00000211933.2 ENSMUST00000212666.2 |
Alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr17_+_36152559 | 0.77 |
ENSMUST00000174124.2
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr6_-_57821483 | 0.76 |
ENSMUST00000226191.2
|
Vmn1r21
|
vomeronasal 1 receptor 21 |
| chr19_-_56996617 | 0.76 |
ENSMUST00000118800.8
ENSMUST00000111584.9 ENSMUST00000122359.8 ENSMUST00000148049.8 |
Afap1l2
|
actin filament associated protein 1-like 2 |
| chr6_-_149090146 | 0.76 |
ENSMUST00000095319.10
ENSMUST00000141346.2 ENSMUST00000111535.8 |
Amn1
|
antagonist of mitotic exit network 1 |
| chr7_+_129193581 | 0.74 |
ENSMUST00000084519.7
|
Wdr11
|
WD repeat domain 11 |
| chr15_+_12205135 | 0.74 |
ENSMUST00000071993.13
|
Mtmr12
|
myotubularin related protein 12 |
| chr18_+_77981636 | 0.74 |
ENSMUST00000044622.7
|
Epg5
|
ectopic P-granules autophagy protein 5 homolog (C. elegans) |
| chr2_+_164627743 | 0.73 |
ENSMUST00000174070.8
ENSMUST00000172577.8 ENSMUST00000056181.7 |
Snx21
|
sorting nexin family member 21 |
| chrX_-_88976458 | 0.73 |
ENSMUST00000088146.3
|
4930415L06Rik
|
RIKEN cDNA 4930415L06 gene |
| chr7_+_126376099 | 0.72 |
ENSMUST00000038614.12
ENSMUST00000170882.8 ENSMUST00000106359.2 ENSMUST00000106357.8 ENSMUST00000145762.8 |
Ypel3
|
yippee like 3 |
| chr18_+_37646674 | 0.72 |
ENSMUST00000061405.6
|
Pcdhb21
|
protocadherin beta 21 |
| chr8_-_106198112 | 0.71 |
ENSMUST00000014990.13
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chrX_-_20955370 | 0.71 |
ENSMUST00000040667.13
|
Zfp300
|
zinc finger protein 300 |
| chr12_+_102724223 | 0.71 |
ENSMUST00000046404.8
|
Ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 (putative) |
| chr7_-_3551003 | 0.70 |
ENSMUST00000065703.9
ENSMUST00000203020.3 ENSMUST00000203821.3 |
Tarm1
|
T cell-interacting, activating receptor on myeloid cells 1 |
| chr2_+_127050281 | 0.70 |
ENSMUST00000103220.4
|
Snrnp200
|
small nuclear ribonucleoprotein 200 (U5) |
| chr7_+_27869115 | 0.70 |
ENSMUST00000042405.8
|
Fbl
|
fibrillarin |
| chr7_-_29605949 | 0.69 |
ENSMUST00000159920.2
ENSMUST00000162592.8 |
Zfp27
|
zinc finger protein 27 |
| chr5_+_129864044 | 0.68 |
ENSMUST00000201414.5
|
Cct6a
|
chaperonin containing Tcp1, subunit 6a (zeta) |
| chr9_-_36637923 | 0.67 |
ENSMUST00000034625.12
|
Chek1
|
checkpoint kinase 1 |
| chr13_+_38223023 | 0.67 |
ENSMUST00000226110.2
|
Riok1
|
RIO kinase 1 |
| chr7_+_100021425 | 0.66 |
ENSMUST00000098259.11
ENSMUST00000051777.15 |
C2cd3
|
C2 calcium-dependent domain containing 3 |
| chr2_-_164120215 | 0.65 |
ENSMUST00000017142.3
|
Svs4
|
seminal vesicle secretory protein 4 |
| chr7_-_100021514 | 0.65 |
ENSMUST00000032963.10
|
Ppme1
|
protein phosphatase methylesterase 1 |
| chr9_+_26941453 | 0.65 |
ENSMUST00000073127.14
ENSMUST00000086198.5 |
Ncapd3
|
non-SMC condensin II complex, subunit D3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.7 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.7 | 2.0 | GO:0010705 | meiotic DNA double-strand break processing involved in reciprocal meiotic recombination(GO:0010705) |
| 0.6 | 3.1 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.5 | 4.2 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.5 | 1.5 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.5 | 3.4 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.5 | 1.4 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.4 | 3.3 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.4 | 1.6 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 0.4 | 2.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.3 | 4.0 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.3 | 3.6 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.3 | 1.5 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.3 | 3.8 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.3 | 3.4 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.3 | 1.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.3 | 2.5 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.3 | 0.8 | GO:0099578 | regulation of translation at synapse, modulating synaptic transmission(GO:0099547) regulation of translation at postsynapse, modulating synaptic transmission(GO:0099578) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.3 | 0.8 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 0.3 | 0.8 | GO:0033967 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.3 | 2.5 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.2 | 1.2 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.2 | 1.6 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.2 | 1.3 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.2 | 1.8 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 0.6 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.2 | 1.6 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.2 | 0.6 | GO:1904172 | positive regulation of bleb assembly(GO:1904172) |
| 0.2 | 1.9 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.2 | 0.6 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.2 | 0.6 | GO:1901873 | negative regulation of cellular respiration(GO:1901856) regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.2 | 2.2 | GO:0035879 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.2 | 1.8 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.2 | 1.2 | GO:0051105 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.2 | 2.0 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 1.4 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.2 | 1.3 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) |
| 0.2 | 0.5 | GO:0031508 | chromatin remodeling at centromere(GO:0031055) pericentric heterochromatin assembly(GO:0031508) |
| 0.2 | 0.6 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.1 | 1.7 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.1 | 0.9 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 1.6 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 8.1 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.1 | 1.0 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 1.1 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.1 | 1.8 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.1 | 2.5 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 0.5 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.1 | 0.4 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.1 | 2.1 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 1.3 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 2.0 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.7 | GO:0061511 | neural plate axis specification(GO:0021997) centriole elongation(GO:0061511) |
| 0.1 | 0.9 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 1.9 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.1 | 1.6 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.1 | 1.7 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.9 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 1.8 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.8 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 1.8 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 0.5 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.6 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.1 | 1.8 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.1 | 1.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.7 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.1 | 0.4 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 0.2 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.1 | 0.7 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.3 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 1.5 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 1.8 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.1 | 0.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) cap-dependent translational initiation(GO:0002191) |
| 0.1 | 0.7 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 2.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 0.5 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.1 | 3.4 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.1 | 0.6 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.1 | 1.1 | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.1 | 2.4 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 2.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.4 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 1.9 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.1 | 5.1 | GO:1901799 | negative regulation of proteasomal protein catabolic process(GO:1901799) |
| 0.1 | 0.4 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 2.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0048822 | enucleate erythrocyte development(GO:0048822) |
| 0.0 | 1.3 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 2.3 | GO:0051220 | cytoplasmic sequestering of protein(GO:0051220) |
| 0.0 | 1.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 2.3 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 2.1 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 | 0.8 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.9 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.1 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.8 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.8 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.0 | 1.5 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.6 | GO:0070203 | regulation of establishment of protein localization to telomere(GO:0070203) positive regulation of establishment of protein localization to telomere(GO:1904851) |
| 0.0 | 2.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 1.4 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 2.4 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 1.2 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 0.3 | GO:0031038 | myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.0 | 0.3 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.0 | 0.8 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.5 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 4.6 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.6 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.5 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 2.3 | GO:0034620 | cellular response to unfolded protein(GO:0034620) |
| 0.0 | 2.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.5 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 2.2 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.8 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.2 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.8 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.0 | 0.1 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.7 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.2 | GO:0001835 | blastocyst hatching(GO:0001835) DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 2.3 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.4 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.7 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.2 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.2 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.0 | 0.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 1.1 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.0 | 1.4 | GO:0046546 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.4 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 8.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.5 | 4.2 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.4 | 1.6 | GO:0071920 | cleavage body(GO:0071920) |
| 0.4 | 1.8 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.3 | 2.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.3 | 4.7 | GO:0032009 | early phagosome(GO:0032009) |
| 0.2 | 0.8 | GO:0019034 | viral replication complex(GO:0019034) dendritic filopodium(GO:1902737) |
| 0.2 | 2.5 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.2 | 1.4 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.2 | 2.0 | GO:0000801 | central element(GO:0000801) |
| 0.2 | 1.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 1.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 1.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.9 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 2.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 1.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.6 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.6 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 1.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.5 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 3.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.9 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 0.9 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 3.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.8 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 0.4 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 0.8 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 0.9 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 2.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 1.5 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 2.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.9 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.4 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 1.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 3.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 3.4 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.5 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 4.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.7 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 0.6 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.7 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 2.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 3.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.7 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 2.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.6 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.9 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 1.0 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 3.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 3.1 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 4.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 3.9 | GO:0044853 | plasma membrane raft(GO:0044853) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.8 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.5 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.4 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 1.8 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 1.5 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 2.2 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.7 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 1.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.4 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 8.2 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 4.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 8.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.7 | 3.7 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.7 | 4.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.4 | 1.8 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.4 | 1.6 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.4 | 2.0 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.4 | 1.5 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.4 | 1.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.4 | 3.6 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.4 | 2.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.3 | 2.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.3 | 1.9 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.3 | 0.9 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.3 | 2.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.3 | 3.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.3 | 1.6 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.3 | 0.8 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.2 | 2.7 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.2 | 1.7 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.2 | 2.8 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 0.8 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.2 | 2.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 1.0 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 2.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.2 | 1.7 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 2.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) cobalt ion binding(GO:0050897) |
| 0.1 | 0.7 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.1 | 0.5 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 0.4 | GO:0001034 | RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.1 | 0.8 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 2.0 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.8 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 2.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 7.0 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 1.8 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.6 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.1 | 2.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.8 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 2.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.3 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.1 | 0.3 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 1.9 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 3.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.3 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.6 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 1.4 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 4.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.5 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 1.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 3.7 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 1.5 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.1 | 1.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.2 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 3.3 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 4.6 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 2.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 2.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.3 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.1 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 1.6 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.7 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 1.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.7 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.9 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 4.3 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.2 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.4 | GO:0004549 | tRNA-specific ribonuclease activity(GO:0004549) |
| 0.0 | 1.3 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 2.3 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 1.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.6 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 1.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 1.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 2.1 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 1.2 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 3.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 1.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 1.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 3.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 2.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 2.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.9 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.2 | 2.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 6.6 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 1.9 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 1.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 2.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 4.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.5 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 5.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 1.1 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 0.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 0.9 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.1 | 1.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 0.8 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 3.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 0.6 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 1.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 1.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 2.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 2.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.7 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 4.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.7 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 3.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |