Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Sp2
Z-value: 0.96
Transcription factors associated with Sp2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sp2
|
ENSMUSG00000018678.14 | Sp2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sp2 | mm39_v1_chr11_-_96868483_96868537 | 0.06 | 6.3e-01 | Click! |
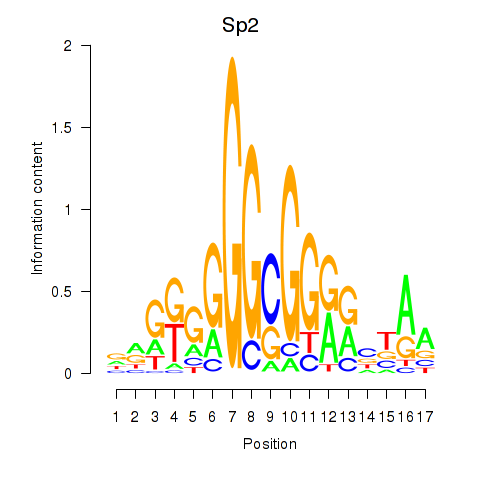
Activity profile of Sp2 motif
Sorted Z-values of Sp2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Sp2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_112586465 | 5.97 |
ENSMUST00000021726.8
|
Adssl1
|
adenylosuccinate synthetase like 1 |
| chr1_+_183766572 | 5.58 |
ENSMUST00000048655.8
|
Dusp10
|
dual specificity phosphatase 10 |
| chr12_+_112586501 | 5.38 |
ENSMUST00000180015.9
|
Adssl1
|
adenylosuccinate synthetase like 1 |
| chr1_-_172047282 | 5.10 |
ENSMUST00000170700.2
ENSMUST00000003554.11 |
Casq1
|
calsequestrin 1 |
| chr1_-_183766195 | 5.02 |
ENSMUST00000050306.8
|
1700056E22Rik
|
RIKEN cDNA 1700056E22 gene |
| chr7_+_19016536 | 3.30 |
ENSMUST00000032559.17
|
Rtn2
|
reticulon 2 (Z-band associated protein) |
| chr14_-_20706556 | 2.85 |
ENSMUST00000090469.8
|
Myoz1
|
myozenin 1 |
| chr7_-_29979758 | 2.80 |
ENSMUST00000108190.8
|
Wdr62
|
WD repeat domain 62 |
| chr7_-_99276310 | 2.79 |
ENSMUST00000178124.3
|
Tpbgl
|
trophoblast glycoprotein-like |
| chr10_-_84938350 | 2.66 |
ENSMUST00000059383.8
ENSMUST00000216889.2 |
Fhl4
|
four and a half LIM domains 4 |
| chr6_-_82751429 | 2.39 |
ENSMUST00000000642.11
|
Hk2
|
hexokinase 2 |
| chr1_-_80318197 | 2.35 |
ENSMUST00000163119.8
|
Cul3
|
cullin 3 |
| chrX_+_149330371 | 2.31 |
ENSMUST00000066337.13
ENSMUST00000112715.2 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr4_-_41045381 | 2.25 |
ENSMUST00000054945.8
|
Aqp7
|
aquaporin 7 |
| chr7_-_24569130 | 2.23 |
ENSMUST00000122995.2
|
Lypd4
|
Ly6/Plaur domain containing 4 |
| chr16_+_4964849 | 2.22 |
ENSMUST00000165810.2
ENSMUST00000230616.2 |
Sec14l5
|
SEC14-like lipid binding 5 |
| chr18_-_35788255 | 2.21 |
ENSMUST00000190196.5
|
Prob1
|
proline rich basic protein 1 |
| chr11_-_69496655 | 2.13 |
ENSMUST00000047889.13
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide |
| chr2_+_119181703 | 2.12 |
ENSMUST00000028780.4
|
Chac1
|
ChaC, cation transport regulator 1 |
| chr1_+_135980508 | 2.09 |
ENSMUST00000112068.10
|
Cacna1s
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr1_+_135980488 | 2.06 |
ENSMUST00000160641.8
|
Cacna1s
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr3_-_107366868 | 1.98 |
ENSMUST00000009617.10
ENSMUST00000238670.2 |
Kcnc4
|
potassium voltage gated channel, Shaw-related subfamily, member 4 |
| chr16_-_10603389 | 1.97 |
ENSMUST00000229866.2
ENSMUST00000038099.6 |
Socs1
|
suppressor of cytokine signaling 1 |
| chr9_-_32255604 | 1.90 |
ENSMUST00000034533.7
|
Kcnj5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr18_-_35795175 | 1.90 |
ENSMUST00000236574.2
ENSMUST00000236971.2 |
Spata24
|
spermatogenesis associated 24 |
| chr9_-_24685572 | 1.90 |
ENSMUST00000052946.12
ENSMUST00000166018.4 |
Tbx20
|
T-box 20 |
| chr6_+_120643323 | 1.89 |
ENSMUST00000112686.8
|
Cecr2
|
CECR2, histone acetyl-lysine reader |
| chr12_-_36206780 | 1.86 |
ENSMUST00000223382.2
ENSMUST00000020856.6 |
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr18_-_35788240 | 1.78 |
ENSMUST00000097619.2
|
Prob1
|
proline rich basic protein 1 |
| chr11_-_68864666 | 1.75 |
ENSMUST00000038644.5
|
Rangrf
|
RAN guanine nucleotide release factor |
| chr9_-_32255556 | 1.74 |
ENSMUST00000214223.2
|
Kcnj5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr7_-_45546081 | 1.73 |
ENSMUST00000120299.4
ENSMUST00000039049.16 |
Syngr4
|
synaptogyrin 4 |
| chr13_+_35925296 | 1.73 |
ENSMUST00000163595.3
|
Cdyl
|
chromodomain protein, Y chromosome-like |
| chr14_-_20844074 | 1.73 |
ENSMUST00000080440.14
ENSMUST00000100837.11 ENSMUST00000071816.7 |
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr9_-_21202353 | 1.69 |
ENSMUST00000086374.8
|
Cdkn2d
|
cyclin dependent kinase inhibitor 2D |
| chr18_-_35795233 | 1.66 |
ENSMUST00000025209.12
ENSMUST00000096573.4 |
Spata24
|
spermatogenesis associated 24 |
| chr5_+_92750898 | 1.64 |
ENSMUST00000200941.2
ENSMUST00000050952.4 |
Stbd1
|
starch binding domain 1 |
| chr15_-_58695379 | 1.63 |
ENSMUST00000072113.6
|
Tmem65
|
transmembrane protein 65 |
| chr7_+_24569472 | 1.63 |
ENSMUST00000011493.6
|
Dmrtc2
|
doublesex and mab-3 related transcription factor like family C2 |
| chr7_-_97228589 | 1.57 |
ENSMUST00000151840.2
ENSMUST00000135998.8 ENSMUST00000144858.8 ENSMUST00000146605.8 ENSMUST00000072725.12 ENSMUST00000138060.3 ENSMUST00000154853.8 ENSMUST00000136757.8 ENSMUST00000124552.3 |
Aamdc
|
adipogenesis associated Mth938 domain containing |
| chr11_-_116472272 | 1.52 |
ENSMUST00000082152.5
|
Ube2o
|
ubiquitin-conjugating enzyme E2O |
| chr7_-_142211203 | 1.51 |
ENSMUST00000097936.9
ENSMUST00000000033.12 |
Igf2
|
insulin-like growth factor 2 |
| chr12_-_110945415 | 1.50 |
ENSMUST00000135131.2
ENSMUST00000043459.13 ENSMUST00000128353.8 |
Ankrd9
|
ankyrin repeat domain 9 |
| chr5_-_24745970 | 1.49 |
ENSMUST00000117900.8
|
Asb10
|
ankyrin repeat and SOCS box-containing 10 |
| chr17_-_29768531 | 1.49 |
ENSMUST00000168339.3
ENSMUST00000114683.10 ENSMUST00000234620.2 |
Tmem217
|
transmembrane protein 217 |
| chr19_-_5475002 | 1.47 |
ENSMUST00000025853.16
|
Drap1
|
Dr1 associated protein 1 (negative cofactor 2 alpha) |
| chr19_+_44994905 | 1.45 |
ENSMUST00000026227.3
|
Twnk
|
twinkle mtDNA helicase |
| chr19_-_5474934 | 1.45 |
ENSMUST00000113674.8
|
Drap1
|
Dr1 associated protein 1 (negative cofactor 2 alpha) |
| chr15_-_63869818 | 1.44 |
ENSMUST00000164532.3
|
Cyrib
|
CYFIP related Rac1 interactor B |
| chr9_-_14411778 | 1.44 |
ENSMUST00000058796.7
|
Kdm4d
|
lysine (K)-specific demethylase 4D |
| chr11_+_78085560 | 1.42 |
ENSMUST00000108317.3
|
Proca1
|
protein interacting with cyclin A1 |
| chr8_+_106554353 | 1.42 |
ENSMUST00000212566.2
ENSMUST00000034365.5 |
Tsnaxip1
|
translin-associated factor X (Tsnax) interacting protein 1 |
| chr13_+_83672654 | 1.41 |
ENSMUST00000199019.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr7_+_45084300 | 1.40 |
ENSMUST00000211150.2
|
Gys1
|
glycogen synthase 1, muscle |
| chr7_+_45084257 | 1.40 |
ENSMUST00000003964.17
|
Gys1
|
glycogen synthase 1, muscle |
| chr11_+_78085522 | 1.40 |
ENSMUST00000060539.13
|
Proca1
|
protein interacting with cyclin A1 |
| chr17_-_29768586 | 1.39 |
ENSMUST00000234305.2
ENSMUST00000234648.2 ENSMUST00000234979.2 |
Gm17657
Tmem217
|
predicted gene, 17657 transmembrane protein 217 |
| chr9_-_66826928 | 1.39 |
ENSMUST00000041139.9
|
Rab8b
|
RAB8B, member RAS oncogene family |
| chr13_+_83672708 | 1.38 |
ENSMUST00000199105.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr10_-_80165257 | 1.37 |
ENSMUST00000020340.15
|
Pcsk4
|
proprotein convertase subtilisin/kexin type 4 |
| chr19_-_5474787 | 1.36 |
ENSMUST00000148219.9
|
Drap1
|
Dr1 associated protein 1 (negative cofactor 2 alpha) |
| chr19_-_5474509 | 1.34 |
ENSMUST00000136579.3
ENSMUST00000113673.9 |
Drap1
|
Dr1 associated protein 1 (negative cofactor 2 alpha) |
| chr19_-_44994824 | 1.34 |
ENSMUST00000097715.4
|
Mrpl43
|
mitochondrial ribosomal protein L43 |
| chrX_+_106860083 | 1.34 |
ENSMUST00000143975.8
ENSMUST00000144695.8 ENSMUST00000167154.2 |
Tent5d
|
terminal nucleotidyltransferase 5D |
| chr17_+_35201074 | 1.34 |
ENSMUST00000007266.14
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr12_-_110945376 | 1.33 |
ENSMUST00000142012.2
|
Ankrd9
|
ankyrin repeat domain 9 |
| chr1_-_155617773 | 1.33 |
ENSMUST00000027740.14
|
Lhx4
|
LIM homeobox protein 4 |
| chr11_-_88609048 | 1.33 |
ENSMUST00000107909.8
|
Msi2
|
musashi RNA-binding protein 2 |
| chr9_-_48747232 | 1.32 |
ENSMUST00000093852.5
|
Zbtb16
|
zinc finger and BTB domain containing 16 |
| chr19_-_53932867 | 1.31 |
ENSMUST00000235688.2
ENSMUST00000235348.2 |
Bbip1
|
BBSome interacting protein 1 |
| chr9_+_21077010 | 1.30 |
ENSMUST00000039413.15
|
Pde4a
|
phosphodiesterase 4A, cAMP specific |
| chr11_-_88608958 | 1.30 |
ENSMUST00000107908.2
|
Msi2
|
musashi RNA-binding protein 2 |
| chr3_+_68912302 | 1.30 |
ENSMUST00000136502.8
ENSMUST00000107803.7 |
Smc4
|
structural maintenance of chromosomes 4 |
| chr17_+_35201130 | 1.30 |
ENSMUST00000173004.2
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr9_-_21202545 | 1.29 |
ENSMUST00000215619.2
|
Cdkn2d
|
cyclin dependent kinase inhibitor 2D |
| chr19_-_41790458 | 1.27 |
ENSMUST00000026150.15
ENSMUST00000163265.9 ENSMUST00000177495.2 |
Arhgap19
|
Rho GTPase activating protein 19 |
| chr7_-_28092113 | 1.26 |
ENSMUST00000003536.9
|
Med29
|
mediator complex subunit 29 |
| chr10_-_102864904 | 1.22 |
ENSMUST00000167156.9
|
Alx1
|
ALX homeobox 1 |
| chr4_+_148215339 | 1.22 |
ENSMUST00000084129.9
|
Mad2l2
|
MAD2 mitotic arrest deficient-like 2 |
| chr7_+_28092370 | 1.20 |
ENSMUST00000003529.9
|
Paf1
|
Paf1, RNA polymerase II complex component |
| chr9_-_21202693 | 1.20 |
ENSMUST00000213407.2
|
Cdkn2d
|
cyclin dependent kinase inhibitor 2D |
| chr17_+_25023263 | 1.19 |
ENSMUST00000234372.2
ENSMUST00000024972.7 |
Meiob
|
meiosis specific with OB domains |
| chr10_+_80003612 | 1.19 |
ENSMUST00000105365.9
|
Cirbp
|
cold inducible RNA binding protein |
| chr6_+_42326528 | 1.19 |
ENSMUST00000203329.3
|
Zyx
|
zyxin |
| chr12_-_36206626 | 1.18 |
ENSMUST00000220828.2
|
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr9_-_14411690 | 1.18 |
ENSMUST00000115647.3
|
Kdm4d
|
lysine (K)-specific demethylase 4D |
| chr12_-_36206750 | 1.16 |
ENSMUST00000221388.2
|
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr18_+_64473091 | 1.15 |
ENSMUST00000175965.10
|
Onecut2
|
one cut domain, family member 2 |
| chr7_+_101467512 | 1.15 |
ENSMUST00000008090.11
|
Phox2a
|
paired-like homeobox 2a |
| chr10_-_102864921 | 1.15 |
ENSMUST00000217946.2
ENSMUST00000219194.2 |
Alx1
|
ALX homeobox 1 |
| chr7_-_24371457 | 1.15 |
ENSMUST00000078001.7
|
Tex101
|
testis expressed gene 101 |
| chr1_-_74544946 | 1.14 |
ENSMUST00000044260.11
ENSMUST00000186282.7 |
Usp37
|
ubiquitin specific peptidase 37 |
| chr3_+_68912043 | 1.13 |
ENSMUST00000042901.15
|
Smc4
|
structural maintenance of chromosomes 4 |
| chr14_-_63482668 | 1.13 |
ENSMUST00000118022.8
|
Gata4
|
GATA binding protein 4 |
| chr1_+_74545203 | 1.12 |
ENSMUST00000087215.7
|
Cnot9
|
CCR4-NOT transcription complex, subunit 9 |
| chr11_+_69871952 | 1.12 |
ENSMUST00000108593.8
|
Ctdnep1
|
CTD nuclear envelope phosphatase 1 |
| chr9_-_103242737 | 1.11 |
ENSMUST00000072249.13
ENSMUST00000189896.2 |
Cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr14_-_63482720 | 1.11 |
ENSMUST00000067417.10
|
Gata4
|
GATA binding protein 4 |
| chr8_-_114575247 | 1.11 |
ENSMUST00000093113.5
|
Adamts18
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 18 |
| chr9_-_103243039 | 1.11 |
ENSMUST00000035484.11
|
Cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr19_+_6391148 | 1.10 |
ENSMUST00000025897.13
ENSMUST00000130382.8 |
Map4k2
|
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr9_+_40103598 | 1.10 |
ENSMUST00000026693.14
ENSMUST00000168832.2 |
Zfp202
|
zinc finger protein 202 |
| chr18_-_14105670 | 1.08 |
ENSMUST00000025288.9
|
Zfp521
|
zinc finger protein 521 |
| chr6_+_125026865 | 1.06 |
ENSMUST00000112413.8
|
Acrbp
|
proacrosin binding protein |
| chr7_+_45546365 | 1.05 |
ENSMUST00000069772.16
ENSMUST00000210503.2 ENSMUST00000209350.2 |
Tmem143
|
transmembrane protein 143 |
| chr4_-_3938352 | 1.05 |
ENSMUST00000003369.10
|
Plag1
|
pleiomorphic adenoma gene 1 |
| chr11_-_109502243 | 1.05 |
ENSMUST00000103060.10
ENSMUST00000047186.10 ENSMUST00000106689.2 |
Wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr10_+_84412490 | 1.04 |
ENSMUST00000020223.8
|
Tcp11l2
|
t-complex 11 (mouse) like 2 |
| chr11_-_105347500 | 1.04 |
ENSMUST00000049995.10
ENSMUST00000100332.4 |
Marchf10
|
membrane associated ring-CH-type finger 10 |
| chr17_+_56611313 | 1.04 |
ENSMUST00000113035.8
ENSMUST00000113039.9 ENSMUST00000142387.2 |
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr4_-_43040278 | 1.04 |
ENSMUST00000107958.8
ENSMUST00000107959.8 ENSMUST00000152846.8 |
Fam214b
|
family with sequence similarity 214, member B |
| chr1_+_136059101 | 1.03 |
ENSMUST00000075164.11
|
Kif21b
|
kinesin family member 21B |
| chr19_-_14575395 | 1.03 |
ENSMUST00000052011.15
ENSMUST00000167776.3 |
Tle4
|
transducin-like enhancer of split 4 |
| chr7_-_26866157 | 1.03 |
ENSMUST00000080058.11
|
Egln2
|
egl-9 family hypoxia-inducible factor 2 |
| chr15_+_58761057 | 1.02 |
ENSMUST00000036904.7
|
Rnf139
|
ring finger protein 139 |
| chr2_-_35226981 | 1.01 |
ENSMUST00000028241.7
|
Stom
|
stomatin |
| chr3_-_8988854 | 1.01 |
ENSMUST00000042148.6
|
Mrps28
|
mitochondrial ribosomal protein S28 |
| chr10_+_127871493 | 1.00 |
ENSMUST00000092048.13
|
Naca
|
nascent polypeptide-associated complex alpha polypeptide |
| chr11_-_5211558 | 0.99 |
ENSMUST00000020662.15
|
Kremen1
|
kringle containing transmembrane protein 1 |
| chrX_+_160500623 | 0.99 |
ENSMUST00000061514.8
|
Rai2
|
retinoic acid induced 2 |
| chr17_+_29020064 | 0.99 |
ENSMUST00000004985.11
|
Brpf3
|
bromodomain and PHD finger containing, 3 |
| chr5_-_121974913 | 0.99 |
ENSMUST00000040308.14
ENSMUST00000086310.8 |
Sh2b3
|
SH2B adaptor protein 3 |
| chr17_-_35265702 | 0.99 |
ENSMUST00000097338.11
|
Msh5
|
mutS homolog 5 |
| chr10_-_102864632 | 0.99 |
ENSMUST00000040859.6
|
Alx1
|
ALX homeobox 1 |
| chr12_+_112611322 | 0.99 |
ENSMUST00000109755.5
|
Siva1
|
SIVA1, apoptosis-inducing factor |
| chr19_-_5713728 | 0.98 |
ENSMUST00000169854.2
|
Sipa1
|
signal-induced proliferation associated gene 1 |
| chr10_+_127126643 | 0.98 |
ENSMUST00000026475.15
ENSMUST00000139091.2 ENSMUST00000230446.2 |
Ddit3
Ddit3
|
DNA-damage inducible transcript 3 DNA-damage inducible transcript 3 |
| chr3_-_107240989 | 0.98 |
ENSMUST00000061772.11
|
Rbm15
|
RNA binding motif protein 15 |
| chr16_+_91526169 | 0.97 |
ENSMUST00000114001.8
ENSMUST00000113999.8 ENSMUST00000064797.12 ENSMUST00000114002.9 ENSMUST00000095909.10 ENSMUST00000056482.14 ENSMUST00000113996.8 |
Itsn1
|
intersectin 1 (SH3 domain protein 1A) |
| chr19_-_23630143 | 0.97 |
ENSMUST00000035849.5
|
1700028P14Rik
|
RIKEN cDNA 1700028P14 gene |
| chr17_+_35200823 | 0.97 |
ENSMUST00000114011.11
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr19_-_5713648 | 0.96 |
ENSMUST00000080824.13
ENSMUST00000237874.2 ENSMUST00000071857.13 ENSMUST00000236464.2 |
Sipa1
|
signal-induced proliferation associated gene 1 |
| chr13_-_55661240 | 0.96 |
ENSMUST00000069929.13
ENSMUST00000069968.13 ENSMUST00000131306.8 ENSMUST00000046246.13 |
Pdlim7
|
PDZ and LIM domain 7 |
| chr14_+_32321341 | 0.95 |
ENSMUST00000187377.7
ENSMUST00000189022.8 ENSMUST00000186452.7 |
Prrxl1
|
paired related homeobox protein-like 1 |
| chr16_+_91526289 | 0.95 |
ENSMUST00000113993.8
|
Itsn1
|
intersectin 1 (SH3 domain protein 1A) |
| chr10_+_127928622 | 0.93 |
ENSMUST00000219072.2
ENSMUST00000045621.9 ENSMUST00000170054.9 |
Baz2a
|
bromodomain adjacent to zinc finger domain, 2A |
| chr7_-_26865838 | 0.93 |
ENSMUST00000108382.2
|
Egln2
|
egl-9 family hypoxia-inducible factor 2 |
| chr19_+_25482982 | 0.92 |
ENSMUST00000025755.11
|
Dmrt1
|
doublesex and mab-3 related transcription factor 1 |
| chr13_-_58421935 | 0.90 |
ENSMUST00000091579.6
|
Gkap1
|
G kinase anchoring protein 1 |
| chr2_+_102904014 | 0.90 |
ENSMUST00000011055.7
|
Apip
|
APAF1 interacting protein |
| chr2_+_146697746 | 0.89 |
ENSMUST00000099278.9
ENSMUST00000156232.2 |
Kiz
|
kizuna centrosomal protein |
| chr4_-_43045685 | 0.88 |
ENSMUST00000107956.8
ENSMUST00000107957.8 |
Fam214b
|
family with sequence similarity 214, member B |
| chr10_+_128030500 | 0.87 |
ENSMUST00000123291.2
|
Gls2
|
glutaminase 2 (liver, mitochondrial) |
| chr19_+_25483070 | 0.87 |
ENSMUST00000087525.5
|
Dmrt1
|
doublesex and mab-3 related transcription factor 1 |
| chr13_-_41233014 | 0.87 |
ENSMUST00000224423.2
|
Mak
|
male germ cell-associated kinase |
| chr8_-_85696369 | 0.86 |
ENSMUST00000109736.9
ENSMUST00000140561.8 |
Rnaseh2a
|
ribonuclease H2, large subunit |
| chr13_-_58421910 | 0.86 |
ENSMUST00000224505.2
|
Gkap1
|
G kinase anchoring protein 1 |
| chr14_+_55815999 | 0.86 |
ENSMUST00000172738.2
ENSMUST00000089619.13 |
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr19_-_5713701 | 0.85 |
ENSMUST00000164304.9
ENSMUST00000237544.2 |
Sipa1
|
signal-induced proliferation associated gene 1 |
| chr15_+_99192968 | 0.85 |
ENSMUST00000128352.8
ENSMUST00000145482.8 |
Prpf40b
|
pre-mRNA processing factor 40B |
| chr9_+_44893077 | 0.85 |
ENSMUST00000034602.9
|
Cd3d
|
CD3 antigen, delta polypeptide |
| chr1_+_131755715 | 0.85 |
ENSMUST00000086559.7
|
Slc41a1
|
solute carrier family 41, member 1 |
| chr2_-_129139125 | 0.85 |
ENSMUST00000052708.7
|
Ckap2l
|
cytoskeleton associated protein 2-like |
| chr14_-_31552608 | 0.85 |
ENSMUST00000014640.9
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr19_+_8944369 | 0.84 |
ENSMUST00000052248.8
|
Eef1g
|
eukaryotic translation elongation factor 1 gamma |
| chr15_-_72932853 | 0.84 |
ENSMUST00000170633.9
ENSMUST00000228960.2 |
Trappc9
|
trafficking protein particle complex 9 |
| chr7_+_141027557 | 0.84 |
ENSMUST00000106004.8
|
Rplp2
|
ribosomal protein, large P2 |
| chr1_-_93406515 | 0.84 |
ENSMUST00000170883.8
ENSMUST00000189025.7 |
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr1_-_191129223 | 0.83 |
ENSMUST00000067976.9
|
Ppp2r5a
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr12_+_3941728 | 0.83 |
ENSMUST00000172689.8
ENSMUST00000111186.8 |
Dnmt3a
|
DNA methyltransferase 3A |
| chr18_-_58343423 | 0.82 |
ENSMUST00000025497.8
|
Fbn2
|
fibrillin 2 |
| chr10_+_117977708 | 0.82 |
ENSMUST00000020437.13
ENSMUST00000163238.9 |
Mdm1
|
transformed mouse 3T3 cell double minute 1 |
| chr2_+_109111083 | 0.82 |
ENSMUST00000028527.8
|
Kif18a
|
kinesin family member 18A |
| chr4_+_108436639 | 0.81 |
ENSMUST00000102744.4
|
Orc1
|
origin recognition complex, subunit 1 |
| chr18_-_46658957 | 0.80 |
ENSMUST00000036226.6
|
Fem1c
|
fem 1 homolog c |
| chr8_-_85696040 | 0.80 |
ENSMUST00000214133.2
ENSMUST00000147812.8 |
Gm49661
Rnaseh2a
|
predicted gene, 49661 ribonuclease H2, large subunit |
| chr10_+_117977822 | 0.80 |
ENSMUST00000164077.9
ENSMUST00000169817.3 |
Mdm1
|
transformed mouse 3T3 cell double minute 1 |
| chr12_+_112645237 | 0.79 |
ENSMUST00000174780.2
ENSMUST00000169593.2 ENSMUST00000173942.2 |
Zbtb42
|
zinc finger and BTB domain containing 42 |
| chr11_-_96868483 | 0.79 |
ENSMUST00000107624.8
|
Sp2
|
Sp2 transcription factor |
| chr11_-_106051533 | 0.79 |
ENSMUST00000106875.2
|
Limd2
|
LIM domain containing 2 |
| chr7_+_141027763 | 0.78 |
ENSMUST00000106003.2
|
Rplp2
|
ribosomal protein, large P2 |
| chrX_-_72703330 | 0.77 |
ENSMUST00000114473.8
ENSMUST00000002087.14 |
Pnck
|
pregnancy upregulated non-ubiquitously expressed CaM kinase |
| chr12_-_85224062 | 0.77 |
ENSMUST00000004913.7
|
Pgf
|
placental growth factor |
| chr11_+_94102255 | 0.77 |
ENSMUST00000041589.6
|
Tob1
|
transducer of ErbB-2.1 |
| chr8_+_117648474 | 0.77 |
ENSMUST00000034205.5
ENSMUST00000212775.2 |
Cenpn
|
centromere protein N |
| chr7_+_142558837 | 0.76 |
ENSMUST00000207211.2
|
Tspan32
|
tetraspanin 32 |
| chr7_+_101545547 | 0.75 |
ENSMUST00000035395.14
ENSMUST00000106973.8 ENSMUST00000144207.9 |
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr7_+_29979512 | 0.75 |
ENSMUST00000108187.8
ENSMUST00000014072.6 |
1700020L13Rik
|
RIKEN cDNA 1700020L13 gene |
| chr4_+_3938903 | 0.74 |
ENSMUST00000121210.8
ENSMUST00000121651.8 ENSMUST00000041122.11 ENSMUST00000120732.8 ENSMUST00000119307.8 ENSMUST00000123769.8 |
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr8_+_111821262 | 0.74 |
ENSMUST00000135302.8
ENSMUST00000039333.10 |
Pdpr
|
pyruvate dehydrogenase phosphatase regulatory subunit |
| chr4_+_3938881 | 0.74 |
ENSMUST00000108386.8
ENSMUST00000121110.8 ENSMUST00000149544.8 |
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr11_-_70860778 | 0.74 |
ENSMUST00000108530.2
ENSMUST00000035283.11 ENSMUST00000108531.8 |
Nup88
|
nucleoporin 88 |
| chr14_+_32321824 | 0.74 |
ENSMUST00000068938.7
ENSMUST00000228878.2 |
Prrxl1
|
paired related homeobox protein-like 1 |
| chr2_-_102903680 | 0.73 |
ENSMUST00000132449.8
ENSMUST00000111183.2 ENSMUST00000011058.9 |
Pdhx
|
pyruvate dehydrogenase complex, component X |
| chr13_-_41233104 | 0.73 |
ENSMUST00000070193.14
ENSMUST00000165087.8 |
Mak
|
male germ cell-associated kinase |
| chr2_+_164647002 | 0.73 |
ENSMUST00000052107.5
|
Zswim3
|
zinc finger SWIM-type containing 3 |
| chr7_-_5017642 | 0.72 |
ENSMUST00000207412.2
ENSMUST00000077385.15 ENSMUST00000165320.3 |
Fiz1
|
Flt3 interacting zinc finger protein 1 |
| chrX_-_52203405 | 0.72 |
ENSMUST00000114843.9
|
Plac1
|
placental specific protein 1 |
| chr11_-_69872050 | 0.71 |
ENSMUST00000108594.8
|
Elp5
|
elongator acetyltransferase complex subunit 5 |
| chr10_-_44334711 | 0.71 |
ENSMUST00000039174.11
|
Prdm1
|
PR domain containing 1, with ZNF domain |
| chr8_-_85567256 | 0.71 |
ENSMUST00000003911.13
ENSMUST00000109761.9 ENSMUST00000128035.2 |
Rad23a
|
RAD23 homolog A, nucleotide excision repair protein |
| chr5_-_121974873 | 0.71 |
ENSMUST00000118580.6
|
Sh2b3
|
SH2B adaptor protein 3 |
| chr2_+_104956850 | 0.70 |
ENSMUST00000143043.8
|
Wt1
|
Wilms tumor 1 homolog |
| chr1_-_189901596 | 0.70 |
ENSMUST00000010319.14
|
Prox1
|
prospero homeobox 1 |
| chr14_+_121148625 | 0.70 |
ENSMUST00000032898.9
|
Ipo5
|
importin 5 |
| chr2_+_119803230 | 0.70 |
ENSMUST00000229024.2
|
Mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr7_+_27147403 | 0.69 |
ENSMUST00000037399.16
ENSMUST00000108358.8 |
Blvrb
|
biliverdin reductase B (flavin reductase (NADPH)) |
| chr1_-_160862364 | 0.69 |
ENSMUST00000177003.2
ENSMUST00000159250.9 ENSMUST00000162226.9 |
Zbtb37
|
zinc finger and BTB domain containing 37 |
| chr3_+_129007599 | 0.69 |
ENSMUST00000042587.12
|
Pitx2
|
paired-like homeodomain transcription factor 2 |
| chr14_-_25769457 | 0.69 |
ENSMUST00000069180.8
|
Zcchc24
|
zinc finger, CCHC domain containing 24 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 11.3 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 1.3 | 5.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.9 | 2.6 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.8 | 5.6 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.7 | 2.2 | GO:0060540 | lung lobe formation(GO:0060464) diaphragm morphogenesis(GO:0060540) |
| 0.7 | 2.0 | GO:1904456 | negative regulation of neuronal action potential(GO:1904456) |
| 0.6 | 1.9 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.6 | 2.3 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.6 | 2.8 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.5 | 1.6 | GO:0061723 | glycophagy(GO:0061723) |
| 0.5 | 4.8 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.5 | 2.4 | GO:0035795 | negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.4 | 2.2 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.4 | 2.1 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.4 | 1.1 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.4 | 1.1 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.3 | 1.0 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.3 | 2.4 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.3 | 1.3 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.3 | 4.2 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.3 | 2.9 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.3 | 3.9 | GO:0035983 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.3 | 3.6 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.3 | 1.7 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.3 | 0.8 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.3 | 2.1 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.3 | 0.3 | GO:0001743 | optic placode formation(GO:0001743) |
| 0.2 | 2.0 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 0.2 | 1.0 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.2 | 1.2 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.2 | 0.7 | GO:2001074 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 0.2 | 2.5 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.2 | 0.9 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.2 | 0.7 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.2 | 1.8 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.2 | 2.0 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.2 | 1.9 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.2 | 1.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.2 | 0.4 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 0.2 | 0.8 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.2 | 1.7 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.2 | 0.8 | GO:1900110 | negative regulation of histone H3-K9 dimethylation(GO:1900110) |
| 0.2 | 1.1 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.2 | 1.5 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.2 | 0.5 | GO:0097037 | heme export(GO:0097037) |
| 0.2 | 1.3 | GO:1900425 | activation of JNKK activity(GO:0007256) negative regulation of defense response to bacterium(GO:1900425) |
| 0.2 | 1.8 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.2 | 0.9 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.2 | 0.8 | GO:0071692 | protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.2 | 0.8 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.2 | 1.6 | GO:1902856 | negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.2 | 1.5 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.2 | 1.2 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.2 | 2.7 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.2 | 2.1 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 0.7 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.1 | 0.7 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.1 | 0.8 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.1 | 1.9 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.1 | 3.4 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.1 | 0.7 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.1 | 1.6 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 1.5 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 | 1.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.4 | GO:0097402 | neuroblast migration(GO:0097402) |
| 0.1 | 0.9 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.1 | 0.6 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 | 0.7 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 0.9 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 1.5 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 1.8 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 | 0.8 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 1.9 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.1 | 0.3 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.1 | 0.5 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 1.0 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.1 | 0.3 | GO:0035037 | sperm entry(GO:0035037) |
| 0.1 | 1.7 | GO:0021559 | trigeminal nerve development(GO:0021559) |
| 0.1 | 0.3 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.1 | 0.5 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 0.7 | GO:2000049 | regulation of cell-cell adhesion mediated by cadherin(GO:2000047) positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.1 | 0.5 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.1 | 0.8 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 | 0.6 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) regulation of establishment of T cell polarity(GO:1903903) |
| 0.1 | 0.2 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.1 | 0.7 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.1 | 0.5 | GO:0072181 | mesonephric duct formation(GO:0072181) |
| 0.1 | 0.4 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.1 | 2.4 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.1 | 0.8 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 0.4 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.1 | 0.7 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 2.4 | GO:0031442 | positive regulation of mRNA 3'-end processing(GO:0031442) |
| 0.1 | 0.8 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.1 | 0.7 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.8 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 0.5 | GO:0002351 | serotonin production involved in inflammatory response(GO:0002351) serotonin secretion involved in inflammatory response(GO:0002442) serotonin secretion by platelet(GO:0002554) |
| 0.1 | 1.0 | GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901030) |
| 0.1 | 0.4 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 0.7 | GO:0000056 | ribosomal large subunit export from nucleus(GO:0000055) ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.7 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 0.5 | GO:0051410 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.1 | 1.0 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.6 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 0.5 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.9 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.1 | 0.5 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 0.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 | 1.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 1.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 1.0 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.1 | 0.7 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.2 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.1 | 1.4 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.1 | 0.4 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.1 | 2.8 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.3 | GO:0010958 | regulation of amino acid import(GO:0010958) |
| 0.1 | 0.3 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.1 | 0.6 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.6 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.1 | 0.3 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 | 1.2 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.3 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 1.1 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.1 | 1.0 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.1 | 3.4 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.1 | 1.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 0.8 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.1 | 1.0 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.1 | 0.4 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.5 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 1.6 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.1 | 1.4 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 0.8 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.1 | 0.5 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.1 | 0.2 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.1 | 0.8 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 1.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.3 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.1 | 2.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 0.5 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.8 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.6 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.5 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.3 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 2.7 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.4 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 3.1 | GO:0065002 | intracellular protein transmembrane transport(GO:0065002) protein transmembrane transport(GO:0071806) |
| 0.0 | 0.5 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 2.8 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.1 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.0 | 0.3 | GO:0042997 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.2 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.8 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.0 | 0.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.5 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 1.0 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 | 0.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 1.3 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 | 0.7 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.3 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.6 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.2 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.4 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.5 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.4 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.2 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 1.1 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.3 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.8 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.2 | GO:0060452 | negative regulation of phospholipase activity(GO:0010519) positive regulation of cardiac muscle contraction(GO:0060452) regulation of adrenergic receptor signaling pathway(GO:0071877) |
| 0.0 | 0.1 | GO:0001844 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:0001844) |
| 0.0 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.8 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.2 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.4 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.3 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 1.5 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.3 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.1 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.0 | 2.9 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.3 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.5 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.5 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.2 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.3 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.5 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.0 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.0 | 0.8 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.0 | 0.4 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) |
| 0.0 | 0.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.4 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.2 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.8 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.3 | GO:2000678 | negative regulation of transcription regulatory region DNA binding(GO:2000678) |
| 0.0 | 0.2 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.5 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.4 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.4 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.6 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.6 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.7 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.2 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.3 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.4 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 0.4 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 2.0 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 1.2 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.0 | 0.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.1 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.0 | 1.0 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.5 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 1.0 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.5 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.6 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.3 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.0 | 0.3 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.3 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 1.4 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 2.1 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 1.2 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.1 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 1.0 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.0 | 0.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.0 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.4 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.6 | 1.9 | GO:0090537 | CERF complex(GO:0090537) |
| 0.5 | 4.2 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.4 | 3.6 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.4 | 1.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.3 | 2.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.3 | 1.0 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) CHOP-ATF3 complex(GO:1990622) |
| 0.3 | 1.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.3 | 2.4 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.3 | 1.7 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.2 | 2.9 | GO:0000796 | condensin complex(GO:0000796) |
| 0.2 | 0.7 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.2 | 0.6 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.2 | 4.6 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 1.6 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.2 | 0.9 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 0.8 | GO:0070442 | integrin alphaIIb-beta3 complex(GO:0070442) |
| 0.2 | 0.6 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 1.8 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 1.6 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 0.5 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 1.0 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.3 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 0.6 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 0.7 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 1.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.5 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.1 | 0.5 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 0.7 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 0.8 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 1.0 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 1.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 1.3 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.8 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 2.7 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 0.8 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.4 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 1.1 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 2.5 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 0.3 | GO:0000938 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 0.1 | 0.2 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.1 | 1.0 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.5 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 5.6 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.6 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.8 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 1.0 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.5 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.4 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.8 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 6.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.2 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.3 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.2 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.4 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 2.8 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.5 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.0 | 0.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.2 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.3 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.7 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.7 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 1.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 1.0 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 1.6 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 12.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.8 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 1.5 | GO:0034358 | plasma lipoprotein particle(GO:0034358) lipoprotein particle(GO:1990777) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 1.7 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 5.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.1 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 2.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.4 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 1.0 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 2.3 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.0 | 1.4 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 7.4 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 2.1 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 2.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 1.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.9 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.2 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.5 | GO:0030286 | dynein complex(GO:0030286) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.3 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.9 | 2.8 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.8 | 2.3 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.6 | 2.4 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.6 | 2.9 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.5 | 3.6 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.5 | 2.3 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.4 | 1.2 | GO:0042602 | riboflavin reductase (NADPH) activity(GO:0042602) |
| 0.4 | 2.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.3 | 6.0 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.3 | 1.6 | GO:2001070 | glycogen binding(GO:2001069) starch binding(GO:2001070) |
| 0.3 | 1.0 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.3 | 2.4 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 1.7 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.2 | 1.7 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 4.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 2.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.2 | 1.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 0.7 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.2 | 1.8 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.2 | 0.5 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.2 | 5.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 2.0 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.2 | 1.8 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 4.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 3.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.5 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 1.6 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 0.5 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) U7 snRNA binding(GO:0071209) |
| 0.1 | 0.7 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 1.2 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 1.5 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 0.3 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 1.8 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 1.7 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 0.8 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 1.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.5 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.1 | 0.9 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.3 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.1 | 0.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.6 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.4 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 2.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.5 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.3 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 2.8 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 0.5 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.8 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 3.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 3.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 3.0 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.6 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 1.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.4 | GO:1990269 | RNA polymerase II C-terminal domain phosphoserine binding(GO:1990269) |
| 0.1 | 0.5 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.5 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 4.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.6 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.4 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 0.9 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.5 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.8 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.7 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.2 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 1.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.5 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.4 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.3 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 1.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 2.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.3 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.5 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.0 | 0.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 1.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 2.0 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 1.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.0 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.7 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 1.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 1.5 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.4 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 3.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.8 | GO:0015491 | cation:cation antiporter activity(GO:0015491) |
| 0.0 | 0.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 1.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 1.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.7 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 1.7 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.6 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0035033 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 2.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 1.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 2.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.4 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 3.0 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.5 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 2.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 2.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 2.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.5 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 8.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 4.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 2.9 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 5.6 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 4.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 5.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 0.8 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 1.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 2.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.3 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 1.9 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 2.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.2 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 2.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.7 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 2.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.4 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 2.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.6 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 1.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.1 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.2 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.3 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 1.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 3.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 11.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 0.6 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.2 | 4.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.2 | 4.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 2.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 3.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 2.5 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 2.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 1.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 1.0 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 1.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 4.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 3.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.7 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 2.9 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 3.3 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 0.6 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.9 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.5 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 2.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.3 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR IN DISEASE | Genes involved in Signaling by FGFR in disease |
| 0.0 | 0.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 2.8 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.4 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 1.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.8 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 2.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.7 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.5 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 6.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 2.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 2.0 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 2.4 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 1.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |