Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
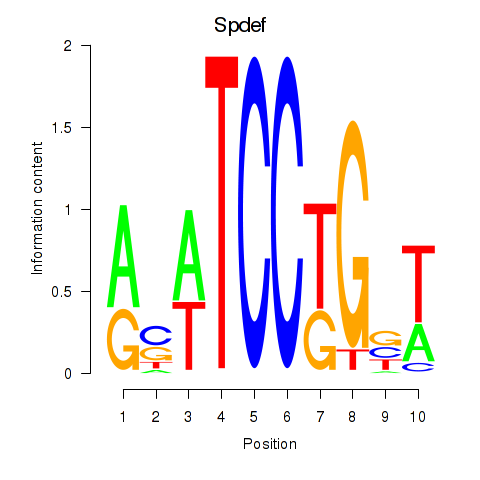
Results for Spdef
Z-value: 1.04
Transcription factors associated with Spdef
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Spdef
|
ENSMUSG00000024215.15 | Spdef |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Spdef | mm39_v1_chr17_-_27947863_27947930 | 0.72 | 6.8e-13 | Click! |
Activity profile of Spdef motif
Sorted Z-values of Spdef motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Spdef
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_4825216 | 9.21 |
ENSMUST00000185147.8
|
Smim22
|
small integral membrane protein 22 |
| chr7_+_27770655 | 8.83 |
ENSMUST00000138392.8
ENSMUST00000076648.8 |
Fcgbp
|
Fc fragment of IgG binding protein |
| chr11_+_69856222 | 8.04 |
ENSMUST00000018713.13
|
Cldn7
|
claudin 7 |
| chr16_+_4825170 | 7.89 |
ENSMUST00000178155.9
|
Smim22
|
small integral membrane protein 22 |
| chr16_+_4825146 | 7.78 |
ENSMUST00000184439.8
|
Smim22
|
small integral membrane protein 22 |
| chr7_+_130633776 | 7.74 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr3_-_90150393 | 7.21 |
ENSMUST00000107369.2
|
Creb3l4
|
cAMP responsive element binding protein 3-like 4 |
| chr2_-_119060366 | 7.09 |
ENSMUST00000076084.6
|
Ppp1r14d
|
protein phosphatase 1, regulatory inhibitor subunit 14D |
| chr1_+_135746330 | 6.94 |
ENSMUST00000038760.10
|
Lad1
|
ladinin |
| chr8_-_65582206 | 6.88 |
ENSMUST00000098713.5
|
Smim31
|
small integral membrane protein 31 |
| chr2_-_119060330 | 6.65 |
ENSMUST00000110820.3
|
Ppp1r14d
|
protein phosphatase 1, regulatory inhibitor subunit 14D |
| chr10_+_34359513 | 6.57 |
ENSMUST00000170771.3
|
Frk
|
fyn-related kinase |
| chr6_-_86770504 | 6.40 |
ENSMUST00000204441.3
ENSMUST00000204398.2 ENSMUST00000001187.15 |
Anxa4
|
annexin A4 |
| chr10_+_34359395 | 6.33 |
ENSMUST00000019913.15
|
Frk
|
fyn-related kinase |
| chr9_+_44966464 | 5.93 |
ENSMUST00000114664.8
|
Mpzl3
|
myelin protein zero-like 3 |
| chr7_+_19699291 | 5.54 |
ENSMUST00000094753.6
|
Ceacam20
|
carcinoembryonic antigen-related cell adhesion molecule 20 |
| chr19_-_11058452 | 5.33 |
ENSMUST00000025636.8
|
Ms4a8a
|
membrane-spanning 4-domains, subfamily A, member 8A |
| chr4_+_118384426 | 5.29 |
ENSMUST00000030261.6
|
2610528J11Rik
|
RIKEN cDNA 2610528J11 gene |
| chr10_-_79369584 | 5.02 |
ENSMUST00000218241.2
ENSMUST00000166804.2 ENSMUST00000063879.13 |
Plpp2
|
phospholipid phosphatase 2 |
| chr3_-_30194559 | 4.89 |
ENSMUST00000108271.10
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr7_+_140659672 | 4.82 |
ENSMUST00000066873.5
ENSMUST00000163041.2 |
Pkp3
|
plakophilin 3 |
| chr4_+_80828883 | 4.64 |
ENSMUST00000055922.4
|
Lurap1l
|
leucine rich adaptor protein 1-like |
| chr16_-_21814190 | 4.63 |
ENSMUST00000231766.2
ENSMUST00000074230.12 |
Liph
|
lipase, member H |
| chr2_+_129854256 | 4.50 |
ENSMUST00000110299.3
|
Tgm3
|
transglutaminase 3, E polypeptide |
| chr7_+_143027473 | 4.44 |
ENSMUST00000052348.12
|
Slc22a18
|
solute carrier family 22 (organic cation transporter), member 18 |
| chr4_-_130172998 | 4.31 |
ENSMUST00000120126.9
|
Serinc2
|
serine incorporator 2 |
| chr3_-_84489923 | 4.29 |
ENSMUST00000143514.3
|
Arfip1
|
ADP-ribosylation factor interacting protein 1 |
| chr15_-_99717956 | 4.23 |
ENSMUST00000109024.9
|
Lima1
|
LIM domain and actin binding 1 |
| chr4_+_118384183 | 4.17 |
ENSMUST00000106367.8
|
2610528J11Rik
|
RIKEN cDNA 2610528J11 gene |
| chr3_+_27992787 | 4.10 |
ENSMUST00000120834.8
|
Pld1
|
phospholipase D1 |
| chr6_-_72367642 | 4.01 |
ENSMUST00000059983.10
|
Vamp8
|
vesicle-associated membrane protein 8 |
| chr9_-_50657800 | 3.95 |
ENSMUST00000239417.2
ENSMUST00000034564.4 |
2310030G06Rik
|
RIKEN cDNA 2310030G06 gene |
| chr16_-_21814289 | 3.95 |
ENSMUST00000060673.8
|
Liph
|
lipase, member H |
| chr7_-_25315299 | 3.90 |
ENSMUST00000098663.4
ENSMUST00000238895.2 |
Erich4
|
glutamate rich 4 |
| chr17_+_32755525 | 3.70 |
ENSMUST00000169591.8
ENSMUST00000003416.15 |
Cyp4f16
|
cytochrome P450, family 4, subfamily f, polypeptide 16 |
| chr15_-_34679321 | 3.42 |
ENSMUST00000040791.9
|
Nipal2
|
NIPA-like domain containing 2 |
| chr1_+_58841650 | 3.34 |
ENSMUST00000165549.8
|
Casp8
|
caspase 8 |
| chr1_+_58841808 | 3.23 |
ENSMUST00000190213.2
|
Casp8
|
caspase 8 |
| chr15_+_78912642 | 3.23 |
ENSMUST00000180086.3
|
H1f0
|
H1.0 linker histone |
| chr6_-_87327885 | 3.19 |
ENSMUST00000032129.3
|
Gkn1
|
gastrokine 1 |
| chr13_-_58532990 | 3.16 |
ENSMUST00000022032.7
|
2210016F16Rik
|
RIKEN cDNA 2210016F16 gene |
| chr11_-_105835238 | 3.14 |
ENSMUST00000019734.11
ENSMUST00000184269.3 ENSMUST00000150563.3 |
Cyb561
|
cytochrome b-561 |
| chr2_-_120144622 | 3.10 |
ENSMUST00000054651.8
|
Pla2g4f
|
phospholipase A2, group IVF |
| chr17_+_35268942 | 2.98 |
ENSMUST00000007257.10
|
Clic1
|
chloride intracellular channel 1 |
| chr8_+_72914966 | 2.96 |
ENSMUST00000003121.9
|
Rab8a
|
RAB8A, member RAS oncogene family |
| chr2_-_92290791 | 2.95 |
ENSMUST00000125276.2
|
Slc35c1
|
solute carrier family 35, member C1 |
| chr2_+_57887896 | 2.94 |
ENSMUST00000112616.8
ENSMUST00000166729.2 |
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr3_-_84489783 | 2.88 |
ENSMUST00000107687.9
ENSMUST00000098990.10 |
Arfip1
|
ADP-ribosylation factor interacting protein 1 |
| chr17_-_56312555 | 2.87 |
ENSMUST00000043785.8
|
Stap2
|
signal transducing adaptor family member 2 |
| chr14_+_55909816 | 2.84 |
ENSMUST00000227178.2
ENSMUST00000227914.2 |
Gmpr2
|
guanosine monophosphate reductase 2 |
| chr7_-_30672747 | 2.80 |
ENSMUST00000205961.2
|
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr12_-_114579938 | 2.76 |
ENSMUST00000195469.6
ENSMUST00000109711.4 |
Ighv1-12
|
immunoglobulin heavy variable V1-12 |
| chr14_-_54491365 | 2.66 |
ENSMUST00000128231.2
|
Dad1
|
defender against cell death 1 |
| chr18_+_35860019 | 2.65 |
ENSMUST00000097617.3
|
1700066B19Rik
|
RIKEN cDNA 1700066B19 gene |
| chr3_-_89300936 | 2.62 |
ENSMUST00000124783.8
ENSMUST00000126027.8 |
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr18_+_67338437 | 2.62 |
ENSMUST00000210564.3
|
Chmp1b
|
charged multivesicular body protein 1B |
| chr14_-_55344004 | 2.59 |
ENSMUST00000036041.15
|
Ap1g2
|
adaptor protein complex AP-1, gamma 2 subunit |
| chr14_+_55909692 | 2.58 |
ENSMUST00000002397.7
|
Gmpr2
|
guanosine monophosphate reductase 2 |
| chr3_+_90448453 | 2.54 |
ENSMUST00000107333.8
ENSMUST00000107331.8 ENSMUST00000098910.3 |
S100a16
|
S100 calcium binding protein A16 |
| chr16_-_56533179 | 2.53 |
ENSMUST00000136394.8
|
Tfg
|
Trk-fused gene |
| chr19_-_38032006 | 2.50 |
ENSMUST00000172095.3
ENSMUST00000041475.16 |
Myof
|
myoferlin |
| chr12_+_80691275 | 2.50 |
ENSMUST00000217889.2
|
Slc39a9
|
solute carrier family 39 (zinc transporter), member 9 |
| chr5_+_115680964 | 2.50 |
ENSMUST00000137716.8
|
Pxn
|
paxillin |
| chr6_-_119365632 | 2.48 |
ENSMUST00000169744.8
|
Adipor2
|
adiponectin receptor 2 |
| chr17_+_27276262 | 2.44 |
ENSMUST00000049308.9
|
Itpr3
|
inositol 1,4,5-triphosphate receptor 3 |
| chr6_-_95695781 | 2.39 |
ENSMUST00000204224.3
|
Suclg2
|
succinate-Coenzyme A ligase, GDP-forming, beta subunit |
| chr12_+_80690985 | 2.39 |
ENSMUST00000219405.2
ENSMUST00000085245.7 |
Slc39a9
|
solute carrier family 39 (zinc transporter), member 9 |
| chrX_+_36059274 | 2.37 |
ENSMUST00000016463.4
|
Slc25a5
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 5 |
| chr8_+_106032205 | 2.34 |
ENSMUST00000109375.4
ENSMUST00000212033.2 |
Elmo3
|
engulfment and cell motility 3 |
| chr8_+_121854566 | 2.28 |
ENSMUST00000181609.2
|
Foxl1
|
forkhead box L1 |
| chr9_-_22042930 | 2.27 |
ENSMUST00000213815.2
|
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr3_-_108053396 | 2.25 |
ENSMUST00000000001.5
|
Gnai3
|
guanine nucleotide binding protein (G protein), alpha inhibiting 3 |
| chr10_-_128657445 | 2.23 |
ENSMUST00000217685.2
ENSMUST00000026409.5 ENSMUST00000219215.2 ENSMUST00000219524.2 |
Ormdl2
|
ORM1-like 2 (S. cerevisiae) |
| chr13_-_104057016 | 2.22 |
ENSMUST00000022222.12
|
Erbin
|
Erbb2 interacting protein |
| chr1_+_57884693 | 2.20 |
ENSMUST00000169772.3
|
Spats2l
|
spermatogenesis associated, serine-rich 2-like |
| chr19_+_8975249 | 2.20 |
ENSMUST00000236390.2
|
Ahnak
|
AHNAK nucleoprotein (desmoyokin) |
| chr19_-_5156995 | 2.17 |
ENSMUST00000237438.2
ENSMUST00000236149.2 ENSMUST00000025804.7 |
Rab1b
|
RAB1B, member RAS oncogene family |
| chr9_+_80072361 | 2.13 |
ENSMUST00000184480.8
|
Myo6
|
myosin VI |
| chr8_+_72973560 | 2.12 |
ENSMUST00000003123.10
|
Fam32a
|
family with sequence similarity 32, member A |
| chr19_-_38031774 | 2.10 |
ENSMUST00000226068.2
|
Myof
|
myoferlin |
| chr3_-_89300599 | 2.09 |
ENSMUST00000142119.2
ENSMUST00000029677.9 ENSMUST00000148361.8 |
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr12_+_76593799 | 2.06 |
ENSMUST00000218380.2
ENSMUST00000219751.2 |
Plekhg3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr4_-_155141241 | 2.03 |
ENSMUST00000131173.3
|
Plch2
|
phospholipase C, eta 2 |
| chr12_-_114579763 | 1.97 |
ENSMUST00000103500.2
|
Ighv1-12
|
immunoglobulin heavy variable V1-12 |
| chr15_+_94441486 | 1.95 |
ENSMUST00000074936.10
|
Irak4
|
interleukin-1 receptor-associated kinase 4 |
| chr3_-_5641171 | 1.92 |
ENSMUST00000071280.8
ENSMUST00000195855.6 ENSMUST00000165309.8 ENSMUST00000164828.8 |
Pex2
|
peroxisomal biogenesis factor 2 |
| chr7_-_79882228 | 1.91 |
ENSMUST00000123279.8
|
Cib1
|
calcium and integrin binding 1 (calmyrin) |
| chr7_+_28580847 | 1.90 |
ENSMUST00000066880.6
|
Capn12
|
calpain 12 |
| chr2_-_34262012 | 1.86 |
ENSMUST00000113132.9
ENSMUST00000040638.15 |
Pbx3
|
pre B cell leukemia homeobox 3 |
| chr4_-_107780716 | 1.86 |
ENSMUST00000106719.8
ENSMUST00000106720.9 ENSMUST00000131644.2 ENSMUST00000030345.15 |
Cpt2
|
carnitine palmitoyltransferase 2 |
| chr13_+_41040657 | 1.85 |
ENSMUST00000069958.15
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr11_-_75828551 | 1.82 |
ENSMUST00000121287.8
|
Rph3al
|
rabphilin 3A-like (without C2 domains) |
| chr3_-_5641295 | 1.82 |
ENSMUST00000059021.10
|
Pex2
|
peroxisomal biogenesis factor 2 |
| chr8_+_124059414 | 1.82 |
ENSMUST00000010298.7
|
Spire2
|
spire type actin nucleation factor 2 |
| chr15_-_36609812 | 1.81 |
ENSMUST00000226496.2
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr3_+_95801325 | 1.81 |
ENSMUST00000197081.2
ENSMUST00000056710.10 |
Aph1a
|
aph1 homolog A, gamma secretase subunit |
| chr2_-_52632178 | 1.80 |
ENSMUST00000102759.8
ENSMUST00000127316.8 |
Stam2
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 |
| chr9_+_80072274 | 1.80 |
ENSMUST00000035889.15
ENSMUST00000113268.8 |
Myo6
|
myosin VI |
| chr9_-_110305705 | 1.79 |
ENSMUST00000198164.5
ENSMUST00000068025.13 |
Klhl18
|
kelch-like 18 |
| chrX_-_72759748 | 1.77 |
ENSMUST00000002091.6
|
Bcap31
|
B cell receptor associated protein 31 |
| chr2_-_167334746 | 1.76 |
ENSMUST00000109211.9
ENSMUST00000057627.16 |
Spata2
|
spermatogenesis associated 2 |
| chr11_-_103254257 | 1.76 |
ENSMUST00000092557.6
|
Arhgap27
|
Rho GTPase activating protein 27 |
| chr11_-_69576363 | 1.72 |
ENSMUST00000018896.14
|
Tnfsf13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr17_+_28491085 | 1.68 |
ENSMUST00000169040.3
|
Ppard
|
peroxisome proliferator activator receptor delta |
| chr3_-_79536166 | 1.67 |
ENSMUST00000029386.14
|
Etfdh
|
electron transferring flavoprotein, dehydrogenase |
| chr3_-_14843512 | 1.66 |
ENSMUST00000094365.11
|
Car1
|
carbonic anhydrase 1 |
| chr6_+_14901343 | 1.66 |
ENSMUST00000115477.8
|
Foxp2
|
forkhead box P2 |
| chr7_+_27828855 | 1.65 |
ENSMUST00000059886.12
|
9530053A07Rik
|
RIKEN cDNA 9530053A07 gene |
| chr7_-_79882313 | 1.65 |
ENSMUST00000206084.2
ENSMUST00000205996.2 ENSMUST00000071457.12 |
Cib1
|
calcium and integrin binding 1 (calmyrin) |
| chr11_-_70545424 | 1.64 |
ENSMUST00000108549.2
|
Pfn1
|
profilin 1 |
| chr14_-_55909314 | 1.61 |
ENSMUST00000163750.8
|
Nedd8
|
neural precursor cell expressed, developmentally down-regulated gene 8 |
| chr3_+_95801263 | 1.60 |
ENSMUST00000015894.12
|
Aph1a
|
aph1 homolog A, gamma secretase subunit |
| chr11_+_105866030 | 1.58 |
ENSMUST00000001964.8
|
Ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| chr5_+_115681003 | 1.58 |
ENSMUST00000157050.8
|
Pxn
|
paxillin |
| chr18_+_77861656 | 1.57 |
ENSMUST00000114748.2
|
Atp5a1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1 |
| chr13_-_42000958 | 1.56 |
ENSMUST00000072012.10
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr13_-_60325170 | 1.55 |
ENSMUST00000065086.6
|
Gas1
|
growth arrest specific 1 |
| chr11_-_70545450 | 1.54 |
ENSMUST00000018437.3
|
Pfn1
|
profilin 1 |
| chr11_+_20151406 | 1.53 |
ENSMUST00000020358.12
ENSMUST00000109602.8 ENSMUST00000109601.8 ENSMUST00000163483.2 |
Rab1a
|
RAB1A, member RAS oncogene family |
| chr8_-_84059048 | 1.51 |
ENSMUST00000177594.8
ENSMUST00000053902.4 |
Elmod2
|
ELMO/CED-12 domain containing 2 |
| chr13_-_74882328 | 1.50 |
ENSMUST00000223309.2
|
Cast
|
calpastatin |
| chr8_+_3637785 | 1.46 |
ENSMUST00000171962.3
ENSMUST00000207712.2 ENSMUST00000207970.2 ENSMUST00000207533.2 ENSMUST00000208240.2 ENSMUST00000207432.2 ENSMUST00000207077.2 |
Camsap3
|
calmodulin regulated spectrin-associated protein family, member 3 |
| chr13_-_74956924 | 1.45 |
ENSMUST00000223206.2
|
Cast
|
calpastatin |
| chr14_+_28233301 | 1.44 |
ENSMUST00000112272.2
|
Wnt5a
|
wingless-type MMTV integration site family, member 5A |
| chr2_-_127383305 | 1.41 |
ENSMUST00000103214.3
|
Prom2
|
prominin 2 |
| chr4_+_101504909 | 1.41 |
ENSMUST00000030254.15
|
Leprot
|
leptin receptor overlapping transcript |
| chr17_-_33166362 | 1.40 |
ENSMUST00000234083.2
ENSMUST00000075253.13 |
Cyp4f13
|
cytochrome P450, family 4, subfamily f, polypeptide 13 |
| chr16_+_19578981 | 1.40 |
ENSMUST00000079780.10
ENSMUST00000164397.8 |
B3gnt5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr16_-_43836681 | 1.39 |
ENSMUST00000036174.10
|
Gramd1c
|
GRAM domain containing 1C |
| chr17_-_25394445 | 1.39 |
ENSMUST00000224277.2
|
Percc1
|
proline and glutamate rich with coiled coil 1 |
| chr14_-_55909527 | 1.38 |
ENSMUST00000010520.10
|
Nedd8
|
neural precursor cell expressed, developmentally down-regulated gene 8 |
| chr3_+_85946145 | 1.38 |
ENSMUST00000238331.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr3_-_122778052 | 1.38 |
ENSMUST00000199401.2
ENSMUST00000197314.5 ENSMUST00000197934.5 ENSMUST00000090379.7 |
Usp53
|
ubiquitin specific peptidase 53 |
| chr11_+_72686990 | 1.36 |
ENSMUST00000069395.7
ENSMUST00000172220.8 |
Zzef1
|
zinc finger, ZZ-type with EF hand domain 1 |
| chr13_-_74882374 | 1.36 |
ENSMUST00000220738.2
|
Cast
|
calpastatin |
| chr18_-_46730547 | 1.35 |
ENSMUST00000151189.2
|
Tmed7
|
transmembrane p24 trafficking protein 7 |
| chr11_+_72687080 | 1.35 |
ENSMUST00000207107.2
|
Zzef1
|
zinc finger, ZZ-type with EF hand domain 1 |
| chr18_-_73836810 | 1.34 |
ENSMUST00000025393.14
|
Smad4
|
SMAD family member 4 |
| chr17_+_26895344 | 1.33 |
ENSMUST00000015719.16
|
Atp6v0e
|
ATPase, H+ transporting, lysosomal V0 subunit E |
| chr15_-_78687216 | 1.32 |
ENSMUST00000164826.8
|
Card10
|
caspase recruitment domain family, member 10 |
| chr18_-_46730381 | 1.31 |
ENSMUST00000036030.14
|
Tmed7
|
transmembrane p24 trafficking protein 7 |
| chrX_-_133177717 | 1.30 |
ENSMUST00000087541.12
ENSMUST00000087540.4 |
Trmt2b
|
TRM2 tRNA methyltransferase 2B |
| chr16_+_19578945 | 1.30 |
ENSMUST00000121344.8
|
B3gnt5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr2_+_145627900 | 1.28 |
ENSMUST00000110005.8
ENSMUST00000094480.11 |
Rin2
|
Ras and Rab interactor 2 |
| chrX_+_74460275 | 1.26 |
ENSMUST00000118428.8
ENSMUST00000114074.8 ENSMUST00000133781.8 |
Brcc3
|
BRCA1/BRCA2-containing complex, subunit 3 |
| chr7_+_28140450 | 1.26 |
ENSMUST00000135686.2
|
Gmfg
|
glia maturation factor, gamma |
| chr19_+_5118103 | 1.24 |
ENSMUST00000070630.8
|
Cd248
|
CD248 antigen, endosialin |
| chr16_-_94171853 | 1.23 |
ENSMUST00000113914.8
ENSMUST00000113905.8 |
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr8_-_73324877 | 1.22 |
ENSMUST00000058733.9
ENSMUST00000167290.8 |
Smim7
|
small integral membrane protein 7 |
| chr11_-_86574586 | 1.21 |
ENSMUST00000018315.10
|
Vmp1
|
vacuole membrane protein 1 |
| chr7_-_141241632 | 1.20 |
ENSMUST00000239500.1
|
ENSMUSG00000118661.1
|
mucin 6, gastric |
| chr2_-_127383332 | 1.20 |
ENSMUST00000028855.14
|
Prom2
|
prominin 2 |
| chr6_-_129449739 | 1.19 |
ENSMUST00000112076.9
ENSMUST00000184581.3 |
Clec7a
|
C-type lectin domain family 7, member a |
| chr11_-_75828504 | 1.16 |
ENSMUST00000108420.9
|
Rph3al
|
rabphilin 3A-like (without C2 domains) |
| chr10_+_17598961 | 1.16 |
ENSMUST00000038107.9
ENSMUST00000219558.2 ENSMUST00000218370.2 |
Cited2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr15_-_54141816 | 1.16 |
ENSMUST00000079772.4
|
Tnfrsf11b
|
tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) |
| chr12_+_33004178 | 1.16 |
ENSMUST00000020885.13
|
Sypl
|
synaptophysin-like protein |
| chr8_-_70962972 | 1.15 |
ENSMUST00000140679.8
ENSMUST00000129909.8 ENSMUST00000081940.11 |
Uba52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr16_-_92622972 | 1.15 |
ENSMUST00000023673.14
|
Runx1
|
runt related transcription factor 1 |
| chr2_+_25132941 | 1.14 |
ENSMUST00000114355.2
ENSMUST00000060818.2 |
Rnf208
|
ring finger protein 208 |
| chr19_-_17316906 | 1.13 |
ENSMUST00000169897.2
|
Gcnt1
|
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chr3_+_93052089 | 1.12 |
ENSMUST00000107300.7
ENSMUST00000195515.2 |
Crnn
|
cornulin |
| chr3_+_87826834 | 1.12 |
ENSMUST00000137775.2
|
Mrpl24
|
mitochondrial ribosomal protein L24 |
| chr13_-_21900313 | 1.11 |
ENSMUST00000091756.2
|
H2bc13
|
H2B clustered histone 13 |
| chr3_-_79535966 | 1.11 |
ENSMUST00000120992.8
|
Etfdh
|
electron transferring flavoprotein, dehydrogenase |
| chr1_-_13660027 | 1.10 |
ENSMUST00000027068.11
|
Tram1
|
translocating chain-associating membrane protein 1 |
| chr6_+_29348068 | 1.10 |
ENSMUST00000173216.8
ENSMUST00000173694.5 ENSMUST00000172974.8 ENSMUST00000031779.17 ENSMUST00000090481.14 |
Calu
|
calumenin |
| chr4_-_156312996 | 1.10 |
ENSMUST00000105571.4
|
Plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chrX_+_99669343 | 1.10 |
ENSMUST00000048962.4
|
Kif4
|
kinesin family member 4 |
| chr11_-_70537878 | 1.10 |
ENSMUST00000014750.15
|
Slc25a11
|
solute carrier family 25 (mitochondrial carrier oxoglutarate carrier), member 11 |
| chr3_+_122039274 | 1.09 |
ENSMUST00000178826.8
|
Gclm
|
glutamate-cysteine ligase, modifier subunit |
| chr1_+_151631203 | 1.08 |
ENSMUST00000187951.7
ENSMUST00000191070.2 |
Edem3
|
ER degradation enhancer, mannosidase alpha-like 3 |
| chr5_+_124577952 | 1.08 |
ENSMUST00000059580.11
|
Kmt5a
|
lysine methyltransferase 5A |
| chr19_-_4665509 | 1.08 |
ENSMUST00000053597.3
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr10_-_80156337 | 1.08 |
ENSMUST00000020341.9
|
2310011J03Rik
|
RIKEN cDNA 2310011J03 gene |
| chr7_+_86895851 | 1.07 |
ENSMUST00000032781.14
|
Nox4
|
NADPH oxidase 4 |
| chr11_+_97917520 | 1.07 |
ENSMUST00000092425.11
|
Rpl19
|
ribosomal protein L19 |
| chr11_+_101144533 | 1.07 |
ENSMUST00000007533.15
ENSMUST00000042477.13 ENSMUST00000100414.12 ENSMUST00000107280.11 ENSMUST00000121331.2 |
Vps25
|
vacuolar protein sorting 25 |
| chr4_+_101504938 | 1.06 |
ENSMUST00000106927.2
|
Leprot
|
leptin receptor overlapping transcript |
| chr7_+_28140352 | 1.05 |
ENSMUST00000078845.13
|
Gmfg
|
glia maturation factor, gamma |
| chr11_-_55202969 | 1.05 |
ENSMUST00000108864.2
|
Fat2
|
FAT atypical cadherin 2 |
| chr2_+_6327431 | 1.05 |
ENSMUST00000114937.8
|
Usp6nl
|
USP6 N-terminal like |
| chr15_-_78413780 | 1.02 |
ENSMUST00000229185.2
|
C1qtnf6
|
C1q and tumor necrosis factor related protein 6 |
| chr7_+_3706992 | 1.01 |
ENSMUST00000006496.15
ENSMUST00000108623.8 ENSMUST00000139818.2 ENSMUST00000108625.8 |
Rps9
|
ribosomal protein S9 |
| chr5_-_31065036 | 1.01 |
ENSMUST00000132034.5
ENSMUST00000132253.5 |
Ost4
|
oligosaccharyltransferase complex subunit 4 (non-catalytic) |
| chrX_+_74460234 | 1.01 |
ENSMUST00000033544.14
|
Brcc3
|
BRCA1/BRCA2-containing complex, subunit 3 |
| chrX_-_133177638 | 1.00 |
ENSMUST00000113252.8
|
Trmt2b
|
TRM2 tRNA methyltransferase 2B |
| chr4_-_62326853 | 0.99 |
ENSMUST00000107459.2
ENSMUST00000084525.12 |
Cdc26
|
cell division cycle 26 |
| chr1_+_39406897 | 0.98 |
ENSMUST00000086535.12
|
Rpl31
|
ribosomal protein L31 |
| chr12_+_76118790 | 0.98 |
ENSMUST00000131480.8
|
Syne2
|
spectrin repeat containing, nuclear envelope 2 |
| chr12_+_86129329 | 0.97 |
ENSMUST00000054565.8
ENSMUST00000222821.2 ENSMUST00000222905.2 |
Ift43
|
intraflagellar transport 43 |
| chr17_-_24746804 | 0.97 |
ENSMUST00000176353.8
ENSMUST00000176237.8 |
Traf7
|
TNF receptor-associated factor 7 |
| chr2_+_152511381 | 0.96 |
ENSMUST00000125366.8
ENSMUST00000109825.8 ENSMUST00000089059.9 ENSMUST00000079247.4 |
H13
|
histocompatibility 13 |
| chr18_+_31742565 | 0.95 |
ENSMUST00000164667.2
|
B930094E09Rik
|
RIKEN cDNA B930094E09 gene |
| chr9_+_114517812 | 0.95 |
ENSMUST00000047404.7
|
Dync1li1
|
dynein cytoplasmic 1 light intermediate chain 1 |
| chr1_+_39406979 | 0.93 |
ENSMUST00000178079.8
ENSMUST00000179954.8 |
Rpl31
|
ribosomal protein L31 |
| chr11_+_81926394 | 0.93 |
ENSMUST00000000193.6
|
Ccl2
|
chemokine (C-C motif) ligand 2 |
| chr9_-_113537277 | 0.93 |
ENSMUST00000111861.4
ENSMUST00000035086.13 |
Pdcd6ip
|
programmed cell death 6 interacting protein |
| chr16_-_43800109 | 0.92 |
ENSMUST00000231700.2
|
Zdhhc23
|
zinc finger, DHHC domain containing 23 |
| chr12_+_73631562 | 0.91 |
ENSMUST00000021527.15
|
Prkch
|
protein kinase C, eta |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | GO:0046038 | GMP catabolic process(GO:0046038) |
| 1.2 | 4.7 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 1.0 | 4.0 | GO:0090187 | autophagosome docking(GO:0016240) zymogen granule exocytosis(GO:0070625) positive regulation of pancreatic juice secretion(GO:0090187) |
| 1.0 | 1.0 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.9 | 2.8 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.9 | 0.9 | GO:0090264 | immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) |
| 0.9 | 3.6 | GO:0007113 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.8 | 4.8 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.8 | 2.3 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.7 | 6.4 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.7 | 2.6 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.6 | 4.3 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.6 | 1.8 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.6 | 1.7 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.6 | 2.3 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.6 | 2.3 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.6 | 2.2 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.5 | 1.6 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.5 | 3.4 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.5 | 1.4 | GO:0061347 | chemorepulsion of dopaminergic neuron axon(GO:0036518) cervix development(GO:0060067) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) melanocyte proliferation(GO:0097325) regulation of cell proliferation in midbrain(GO:1904933) |
| 0.5 | 6.6 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.5 | 1.8 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.5 | 1.4 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.4 | 1.3 | GO:2000793 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) nephrogenic mesenchyme development(GO:0072076) cell proliferation involved in heart valve development(GO:2000793) |
| 0.4 | 2.2 | GO:0070425 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.4 | 1.8 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.4 | 1.7 | GO:0036508 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) protein demannosylation(GO:0036507) protein alpha-1,2-demannosylation(GO:0036508) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.4 | 2.4 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.4 | 1.2 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 0.4 | 2.7 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.4 | 2.7 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.4 | 8.1 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.3 | 1.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.3 | 1.7 | GO:0071336 | regulation of hair follicle cell proliferation(GO:0071336) |
| 0.3 | 3.4 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.3 | 4.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.3 | 4.9 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.3 | 6.4 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.3 | 1.2 | GO:0035801 | adrenal cortex development(GO:0035801) adrenal cortex formation(GO:0035802) |
| 0.3 | 2.3 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.3 | 0.8 | GO:2000670 | negative regulation of toll-like receptor 2 signaling pathway(GO:0034136) positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.3 | 2.4 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.3 | 4.6 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.3 | 2.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.3 | 6.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.3 | 3.5 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.2 | 2.7 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.2 | 1.2 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.2 | 2.4 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.2 | 0.9 | GO:0035747 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.2 | 4.3 | GO:2000675 | egg activation(GO:0007343) negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.2 | 0.7 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.2 | 1.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.2 | 3.9 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.2 | 3.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.2 | 1.7 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.2 | 1.5 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.2 | 0.8 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.2 | 1.0 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.2 | 2.9 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.2 | 3.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 0.7 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.2 | 3.0 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.2 | 0.5 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.2 | 2.3 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.2 | 0.8 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.2 | 1.1 | GO:1901228 | positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.2 | 0.6 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.2 | 1.4 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.1 | 0.6 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 0.7 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.1 | 1.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 3.0 | GO:1903540 | neurotransmitter receptor transport to plasma membrane(GO:0098877) neurotransmitter receptor transport to postsynaptic membrane(GO:0098969) establishment of protein localization to postsynaptic membrane(GO:1903540) |
| 0.1 | 1.5 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 0.5 | GO:0032888 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.1 | 4.1 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 0.4 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) establishment of viral latency(GO:0019043) |
| 0.1 | 0.7 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 1.5 | GO:0019068 | virion assembly(GO:0019068) |
| 0.1 | 1.2 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.1 | 0.7 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.1 | 0.6 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.1 | 12.0 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 2.8 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 1.5 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.1 | 3.1 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 | 1.5 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 1.1 | GO:0000022 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 3.0 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 1.9 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.1 | 0.4 | GO:1905077 | negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.1 | 0.4 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.1 | 1.0 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.1 | 1.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 1.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 4.4 | GO:0015893 | drug transport(GO:0015893) |
| 0.1 | 1.6 | GO:0042473 | outer ear morphogenesis(GO:0042473) regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 3.6 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 1.8 | GO:0051900 | regulation of mitochondrial depolarization(GO:0051900) |
| 0.1 | 1.2 | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.1 | 0.4 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 4.4 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 1.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 1.0 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.1 | 3.7 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 0.3 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 1.1 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.1 | 0.6 | GO:0070163 | positive regulation of neutrophil apoptotic process(GO:0033031) adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.1 | 0.4 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.1 | 0.4 | GO:0034627 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 7.2 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 3.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 1.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 4.9 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 1.0 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 1.2 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.1 | 3.7 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.1 | 1.8 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.2 | GO:2000847 | negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.1 | 0.3 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.1 | 0.7 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.1 | 0.7 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.1 | 0.7 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 1.4 | GO:0006691 | leukotriene metabolic process(GO:0006691) |
| 0.0 | 0.8 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 1.1 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 | 3.0 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 3.9 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.3 | GO:0015867 | ATP transport(GO:0015867) |
| 0.0 | 0.2 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 1.3 | GO:0043536 | positive regulation of blood vessel endothelial cell migration(GO:0043536) |
| 0.0 | 0.8 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.5 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 1.0 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.2 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 1.1 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 2.2 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 3.0 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 | 2.2 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.2 | GO:1901355 | circadian regulation of translation(GO:0097167) response to rapamycin(GO:1901355) |
| 0.0 | 0.5 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 0.8 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 1.2 | GO:0002446 | neutrophil mediated immunity(GO:0002446) |
| 0.0 | 5.3 | GO:0042633 | molting cycle(GO:0042303) hair cycle(GO:0042633) |
| 0.0 | 0.4 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.9 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.0 | GO:1904732 | regulation of electron carrier activity(GO:1904732) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.0 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 1.7 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.9 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.0 | 0.6 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.2 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 | 0.7 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 2.6 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 2.7 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.0 | 4.7 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.2 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.3 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 | 9.1 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 0.4 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 5.3 | GO:0035304 | regulation of protein dephosphorylation(GO:0035304) |
| 0.0 | 1.4 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.8 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.0 | 0.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:1990428 | miRNA transport(GO:1990428) |
| 0.0 | 0.7 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 2.2 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.1 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.9 | GO:0045761 | regulation of adenylate cyclase activity(GO:0045761) |
| 0.0 | 0.4 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.4 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.2 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.2 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.3 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.3 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.1 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.5 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 1.3 | 6.6 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 1.0 | 4.8 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.7 | 2.8 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.6 | 7.7 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.6 | 1.8 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.5 | 2.6 | GO:0044393 | microspike(GO:0044393) |
| 0.5 | 2.4 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.4 | 2.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.4 | 3.7 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.4 | 1.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.3 | 2.8 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.3 | 0.9 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.3 | 2.4 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.3 | 4.8 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.3 | 1.3 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.3 | 1.8 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.2 | 8.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 3.0 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.2 | 2.3 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.2 | 2.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 2.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.2 | 0.8 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.2 | 8.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 3.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.2 | 5.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.2 | 1.6 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.2 | 2.0 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 3.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.6 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 1.0 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 3.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 2.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.5 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 0.9 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 2.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 2.8 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 4.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.9 | GO:0044299 | C-fiber(GO:0044299) |
| 0.1 | 8.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 6.5 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.0 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 1.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 1.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 3.0 | GO:0097546 | recycling endosome membrane(GO:0055038) ciliary base(GO:0097546) |
| 0.1 | 0.6 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 2.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 2.2 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 1.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 3.3 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 5.2 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 1.9 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 1.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 3.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 3.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 0.8 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.5 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 0.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.7 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.4 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 3.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 3.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 7.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 4.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 1.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.7 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 1.5 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.9 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 1.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 13.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 2.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.3 | GO:0045252 | dihydrolipoyl dehydrogenase complex(GO:0045240) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 3.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 4.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 4.2 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 4.6 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 2.2 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 1.0 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 1.5 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 9.7 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.4 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 3.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 4.1 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 0.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.9 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 1.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 3.3 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 1.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.3 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.4 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.7 | GO:0035375 | zymogen binding(GO:0035375) |
| 1.8 | 5.4 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 1.6 | 6.6 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 1.0 | 3.1 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 1.0 | 4.1 | GO:0051435 | BH4 domain binding(GO:0051435) |
| 0.9 | 2.7 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.8 | 2.4 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.7 | 3.6 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.7 | 2.8 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.7 | 2.7 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.6 | 4.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.6 | 2.4 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.6 | 1.8 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.6 | 1.7 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.6 | 2.8 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.5 | 4.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.5 | 2.5 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.5 | 1.4 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.5 | 1.8 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.4 | 4.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.4 | 1.6 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.4 | 2.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.4 | 7.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.4 | 1.1 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.3 | 3.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.3 | 1.7 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.3 | 5.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.3 | 1.6 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) bradykinin receptor binding(GO:0031711) |
| 0.3 | 4.0 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.3 | 1.9 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.3 | 0.9 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.3 | 4.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.3 | 1.5 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.3 | 3.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.3 | 1.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.3 | 2.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.3 | 3.9 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.3 | 0.8 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.3 | 2.7 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.3 | 2.4 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.3 | 1.3 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.2 | 13.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.2 | 0.9 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.2 | 0.8 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.2 | 2.7 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.2 | 3.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 0.6 | GO:0051538 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.2 | 2.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 7.2 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.2 | 0.7 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 1.3 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.2 | 1.7 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 2.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 2.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.2 | 0.5 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.1 | 0.9 | GO:0070004 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.6 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 4.4 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.1 | 1.7 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 12.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 2.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 2.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 5.9 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 1.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 0.9 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 2.8 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 1.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 2.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 1.4 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 3.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.0 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 1.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.7 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.6 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.1 | 1.8 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 0.3 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 2.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.3 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.1 | 0.8 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 2.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 7.8 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.1 | 0.7 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.1 | 1.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 0.4 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 2.0 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 0.2 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 1.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.6 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 1.8 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 1.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.4 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 0.8 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 1.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.4 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 1.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.9 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 9.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.6 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.9 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 1.5 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 1.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.8 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 1.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 4.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 2.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.2 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.6 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 1.0 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 3.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 15.4 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 2.7 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 2.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.5 | GO:0016502 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.0 | 1.1 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 3.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 2.4 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 0.1 | GO:0089720 | caspase binding(GO:0089720) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 2.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 3.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 3.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 4.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 10.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 4.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 1.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 5.7 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 1.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 4.8 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.9 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.9 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 2.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.7 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.6 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 7.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.9 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.3 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.3 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.9 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.4 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 1.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 3.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | PID CDC42 PATHWAY | CDC42 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.7 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.3 | 4.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.2 | 3.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 2.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.2 | 3.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.2 | 3.9 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 8.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 4.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 3.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 3.7 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.2 | 3.0 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.2 | 2.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 4.5 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 4.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 3.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.6 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 1.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 3.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 4.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 2.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 4.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 1.7 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 9.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 1.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.9 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 0.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 1.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 0.8 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 1.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.0 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF CYCLIN B | Genes involved in APC/C:Cdc20 mediated degradation of Cyclin B |
| 0.0 | 1.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 2.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 3.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 3.7 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 0.5 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 2.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.5 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.7 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.1 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 1.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 2.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 1.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.5 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.6 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.6 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |