Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Srf
Z-value: 2.68
Transcription factors associated with Srf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Srf
|
ENSMUSG00000015605.7 | Srf |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Srf | mm39_v1_chr17_-_46867083_46867114 | 0.44 | 1.1e-04 | Click! |
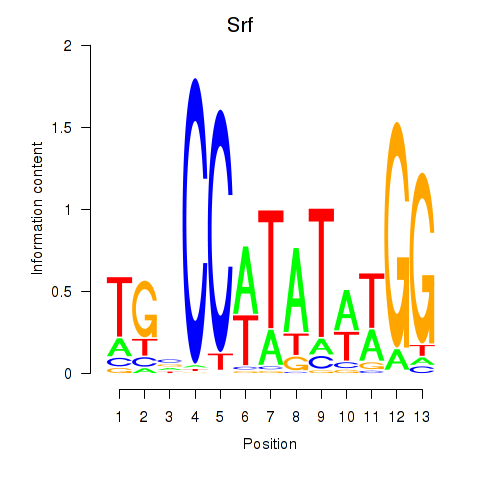
Activity profile of Srf motif
Sorted Z-values of Srf motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Srf
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_113883285 | 33.09 |
ENSMUST00000090269.7
|
Actc1
|
actin, alpha, cardiac muscle 1 |
| chr6_-_83513222 | 32.19 |
ENSMUST00000075161.12
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr6_-_83513184 | 27.92 |
ENSMUST00000205926.2
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr9_+_110592709 | 26.85 |
ENSMUST00000079784.12
|
Myl3
|
myosin, light polypeptide 3 |
| chr12_-_103322226 | 25.74 |
ENSMUST00000021617.14
|
Asb2
|
ankyrin repeat and SOCS box-containing 2 |
| chrX_+_100492684 | 25.23 |
ENSMUST00000033674.6
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr10_+_45211847 | 20.71 |
ENSMUST00000095715.5
|
Bves
|
blood vessel epicardial substance |
| chr8_-_124621483 | 20.69 |
ENSMUST00000034453.6
ENSMUST00000212584.2 |
Acta1
|
actin, alpha 1, skeletal muscle |
| chr1_-_43203051 | 19.56 |
ENSMUST00000008280.14
|
Fhl2
|
four and a half LIM domains 2 |
| chr7_+_19144950 | 19.34 |
ENSMUST00000208710.2
ENSMUST00000003643.3 |
Ckm
|
creatine kinase, muscle |
| chr10_+_32959472 | 18.96 |
ENSMUST00000095762.5
ENSMUST00000218281.2 ENSMUST00000217779.2 ENSMUST00000219665.2 ENSMUST00000219931.2 |
Trdn
|
triadin |
| chr4_+_148085179 | 18.42 |
ENSMUST00000103230.5
|
Nppa
|
natriuretic peptide type A |
| chr8_+_15107646 | 17.65 |
ENSMUST00000033842.4
|
Myom2
|
myomesin 2 |
| chr11_-_5848771 | 17.54 |
ENSMUST00000102921.4
|
Myl7
|
myosin, light polypeptide 7, regulatory |
| chr14_-_34310602 | 17.24 |
ENSMUST00000064098.14
ENSMUST00000090040.12 ENSMUST00000022330.9 ENSMUST00000022327.13 |
Ldb3
|
LIM domain binding 3 |
| chr14_-_55204054 | 15.94 |
ENSMUST00000226297.2
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr14_-_55204092 | 15.78 |
ENSMUST00000081857.14
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr2_+_156617329 | 15.77 |
ENSMUST00000088552.7
|
Myl9
|
myosin, light polypeptide 9, regulatory |
| chr6_+_17306334 | 15.72 |
ENSMUST00000007799.13
ENSMUST00000115456.6 |
Cav1
|
caveolin 1, caveolae protein |
| chr14_-_34310637 | 15.06 |
ENSMUST00000227819.2
|
Ldb3
|
LIM domain binding 3 |
| chr3_+_106020545 | 14.71 |
ENSMUST00000079132.12
ENSMUST00000139086.2 |
Chia1
|
chitinase, acidic 1 |
| chr10_-_120735000 | 14.62 |
ENSMUST00000092143.12
|
Msrb3
|
methionine sulfoxide reductase B3 |
| chr6_-_71239216 | 13.80 |
ENSMUST00000129630.3
ENSMUST00000114186.9 ENSMUST00000074301.10 |
Smyd1
|
SET and MYND domain containing 1 |
| chr8_+_55003359 | 13.45 |
ENSMUST00000033918.4
|
Asb5
|
ankyrin repeat and SOCs box-containing 5 |
| chr14_-_34310438 | 13.37 |
ENSMUST00000228044.2
ENSMUST00000022328.14 |
Ldb3
|
LIM domain binding 3 |
| chr11_+_82802079 | 12.79 |
ENSMUST00000018989.14
ENSMUST00000164945.3 |
Unc45b
|
unc-45 myosin chaperone B |
| chr5_-_113044216 | 11.40 |
ENSMUST00000086617.11
|
Myo18b
|
myosin XVIIIb |
| chr14_-_55204383 | 11.14 |
ENSMUST00000111456.2
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr4_-_141327146 | 10.68 |
ENSMUST00000141518.8
ENSMUST00000127455.8 ENSMUST00000105784.8 |
Fblim1
|
filamin binding LIM protein 1 |
| chr1_-_43235914 | 10.59 |
ENSMUST00000187357.2
|
Fhl2
|
four and a half LIM domains 2 |
| chr14_-_55204023 | 10.26 |
ENSMUST00000124930.8
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr6_+_87350292 | 10.23 |
ENSMUST00000032128.6
|
Gkn2
|
gastrokine 2 |
| chr2_-_60503998 | 9.92 |
ENSMUST00000059888.15
ENSMUST00000154764.2 |
Itgb6
|
integrin beta 6 |
| chr12_+_108389075 | 9.82 |
ENSMUST00000109860.8
|
Eml1
|
echinoderm microtubule associated protein like 1 |
| chr12_-_40249314 | 9.64 |
ENSMUST00000095760.3
|
Lsmem1
|
leucine-rich single-pass membrane protein 1 |
| chr9_-_58220469 | 9.41 |
ENSMUST00000061799.10
|
Loxl1
|
lysyl oxidase-like 1 |
| chr11_-_120622770 | 9.24 |
ENSMUST00000154565.2
ENSMUST00000026148.9 |
Cbr2
|
carbonyl reductase 2 |
| chr11_+_101221431 | 9.24 |
ENSMUST00000103105.10
|
Aoc3
|
amine oxidase, copper containing 3 |
| chr16_-_10614679 | 9.22 |
ENSMUST00000023144.6
|
Prm1
|
protamine 1 |
| chr12_-_40249489 | 9.11 |
ENSMUST00000220951.2
|
Lsmem1
|
leucine-rich single-pass membrane protein 1 |
| chr7_-_142215027 | 8.05 |
ENSMUST00000105936.8
|
Igf2
|
insulin-like growth factor 2 |
| chr5_-_24534554 | 8.01 |
ENSMUST00000115098.7
|
Kcnh2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr1_-_33796790 | 7.75 |
ENSMUST00000187602.2
ENSMUST00000044691.9 |
Bag2
|
BCL2-associated athanogene 2 |
| chr7_+_3341597 | 7.62 |
ENSMUST00000164553.8
|
Myadm
|
myeloid-associated differentiation marker |
| chr16_+_24266829 | 7.36 |
ENSMUST00000078988.10
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr11_-_99383938 | 6.98 |
ENSMUST00000006969.8
|
Krt23
|
keratin 23 |
| chr4_-_141327253 | 6.93 |
ENSMUST00000147785.8
|
Fblim1
|
filamin binding LIM protein 1 |
| chr2_+_150628655 | 6.89 |
ENSMUST00000045441.8
|
Pygb
|
brain glycogen phosphorylase |
| chr4_+_43957401 | 6.72 |
ENSMUST00000030202.14
|
Glipr2
|
GLI pathogenesis-related 2 |
| chr16_+_57369595 | 6.71 |
ENSMUST00000159414.2
|
Filip1l
|
filamin A interacting protein 1-like |
| chr7_+_127845984 | 6.68 |
ENSMUST00000164710.8
ENSMUST00000070656.12 |
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr7_+_127846121 | 6.67 |
ENSMUST00000167965.8
|
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr17_-_73706284 | 6.36 |
ENSMUST00000095208.4
|
Capn13
|
calpain 13 |
| chr3_+_68375495 | 6.31 |
ENSMUST00000182532.8
|
Schip1
|
schwannomin interacting protein 1 |
| chr14_-_45767421 | 6.14 |
ENSMUST00000150660.3
|
Fermt2
|
fermitin family member 2 |
| chr11_+_33913013 | 5.88 |
ENSMUST00000020362.3
|
Kcnmb1
|
potassium large conductance calcium-activated channel, subfamily M, beta member 1 |
| chr11_-_101315345 | 5.30 |
ENSMUST00000107257.8
ENSMUST00000107259.4 ENSMUST00000107252.9 ENSMUST00000093933.11 |
Gm27029
Ptges3l
|
predicted gene, 27029 prostaglandin E synthase 3 like |
| chr2_-_18001734 | 5.24 |
ENSMUST00000105001.4
|
H2al2a
|
H2A histone family member L2A |
| chr4_+_19280850 | 5.03 |
ENSMUST00000102999.2
|
Cngb3
|
cyclic nucleotide gated channel beta 3 |
| chr14_-_34224620 | 4.75 |
ENSMUST00000049005.15
|
Bmpr1a
|
bone morphogenetic protein receptor, type 1A |
| chr1_+_167445815 | 4.63 |
ENSMUST00000111380.2
|
Rxrg
|
retinoid X receptor gamma |
| chr4_+_43957677 | 4.56 |
ENSMUST00000107855.2
|
Glipr2
|
GLI pathogenesis-related 2 |
| chr14_+_20979466 | 4.44 |
ENSMUST00000022369.9
|
Vcl
|
vinculin |
| chr4_+_39450265 | 4.28 |
ENSMUST00000029955.5
|
1700009N14Rik
|
RIKEN cDNA 1700009N14 gene |
| chr12_+_85520652 | 4.16 |
ENSMUST00000021674.7
|
Fos
|
FBJ osteosarcoma oncogene |
| chr17_-_46867083 | 4.05 |
ENSMUST00000015749.7
|
Srf
|
serum response factor |
| chr10_+_4216353 | 4.00 |
ENSMUST00000045730.7
|
Akap12
|
A kinase (PRKA) anchor protein (gravin) 12 |
| chr14_-_34224479 | 3.94 |
ENSMUST00000171551.2
|
Bmpr1a
|
bone morphogenetic protein receptor, type 1A |
| chr10_+_79824418 | 3.49 |
ENSMUST00000004784.11
ENSMUST00000105374.2 |
Cnn2
|
calponin 2 |
| chr15_-_76113692 | 3.46 |
ENSMUST00000074834.12
|
Plec
|
plectin |
| chr17_+_5991555 | 3.38 |
ENSMUST00000115791.10
ENSMUST00000080283.13 |
Synj2
|
synaptojanin 2 |
| chr19_+_53517528 | 3.32 |
ENSMUST00000038287.7
|
Dusp5
|
dual specificity phosphatase 5 |
| chr7_-_44646960 | 3.26 |
ENSMUST00000207443.2
ENSMUST00000207755.2 ENSMUST00000003290.12 |
Bcl2l12
|
BCL2-like 12 (proline rich) |
| chr15_+_102427149 | 3.23 |
ENSMUST00000146756.8
ENSMUST00000142194.3 |
Tarbp2
|
TARBP2, RISC loading complex RNA binding subunit |
| chr1_+_39026887 | 3.14 |
ENSMUST00000194552.2
|
Pdcl3
|
phosducin-like 3 |
| chr14_-_45767575 | 3.06 |
ENSMUST00000045905.15
|
Fermt2
|
fermitin family member 2 |
| chr2_+_20742115 | 3.05 |
ENSMUST00000114606.8
ENSMUST00000114608.3 |
Etl4
|
enhancer trap locus 4 |
| chr14_+_55813074 | 3.00 |
ENSMUST00000022826.7
|
Fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr5_-_100720063 | 2.96 |
ENSMUST00000031264.12
|
Plac8
|
placenta-specific 8 |
| chr1_+_74448535 | 2.90 |
ENSMUST00000027366.13
|
Vil1
|
villin 1 |
| chr3_-_57483330 | 2.87 |
ENSMUST00000120977.2
|
Wwtr1
|
WW domain containing transcription regulator 1 |
| chr11_-_120239301 | 2.83 |
ENSMUST00000062147.14
ENSMUST00000128055.2 |
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr14_-_45767232 | 2.81 |
ENSMUST00000149723.2
|
Fermt2
|
fermitin family member 2 |
| chr6_-_23248361 | 2.76 |
ENSMUST00000031709.7
|
Fezf1
|
Fez family zinc finger 1 |
| chr7_-_142215595 | 2.66 |
ENSMUST00000145896.3
|
Igf2
|
insulin-like growth factor 2 |
| chr18_+_6603627 | 2.63 |
ENSMUST00000234821.2
ENSMUST00000044829.5 ENSMUST00000234337.2 |
4921524L21Rik
|
RIKEN cDNA 4921524L21 gene |
| chr1_+_78794475 | 2.56 |
ENSMUST00000057262.8
ENSMUST00000187432.2 |
Kcne4
|
potassium voltage-gated channel, Isk-related subfamily, gene 4 |
| chr15_+_84926909 | 2.54 |
ENSMUST00000229203.2
|
Fam118a
|
family with sequence similarity 118, member A |
| chr14_+_14901127 | 2.52 |
ENSMUST00000163790.2
|
Gm3558
|
predicted gene 3558 |
| chr14_-_18054325 | 2.43 |
ENSMUST00000168866.2
|
Gm3164
|
predicted gene 3164 |
| chr2_+_69210775 | 2.41 |
ENSMUST00000063690.4
|
Dhrs9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr11_-_120238917 | 2.36 |
ENSMUST00000106215.11
|
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr9_+_108313672 | 2.32 |
ENSMUST00000057265.8
|
BC048562
|
cDNA sequence BC048562 |
| chr2_-_164675357 | 2.30 |
ENSMUST00000042775.5
|
Neurl2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr2_+_154390808 | 2.30 |
ENSMUST00000045116.11
ENSMUST00000109709.4 |
1700003F12Rik
|
RIKEN cDNA 1700003F12 gene |
| chr16_-_92262969 | 2.29 |
ENSMUST00000232239.2
ENSMUST00000060005.15 |
Rcan1
|
regulator of calcineurin 1 |
| chr11_-_120239339 | 2.11 |
ENSMUST00000071555.13
|
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr5_+_16139909 | 2.02 |
ENSMUST00000196750.2
|
Cacna2d1
|
calcium channel, voltage-dependent, alpha2/delta subunit 1 |
| chr11_-_70560110 | 1.83 |
ENSMUST00000129434.2
ENSMUST00000018431.13 |
Spag7
|
sperm associated antigen 7 |
| chr17_-_46464441 | 1.80 |
ENSMUST00000171172.3
|
Mad2l1bp
|
MAD2L1 binding protein |
| chr13_-_21722197 | 1.71 |
ENSMUST00000168629.2
ENSMUST00000218154.2 |
Olfr1366
|
olfactory receptor 1366 |
| chrX_+_11181397 | 1.68 |
ENSMUST00000179004.2
|
H2al1f
|
H2A histone family member L1F |
| chrX_+_11190898 | 1.68 |
ENSMUST00000164729.3
|
H2al1i
|
H2A histone family member L1I |
| chr14_+_16431564 | 1.67 |
ENSMUST00000164139.2
|
Gm8206
|
predicted gene 8206 |
| chr14_-_16968099 | 1.64 |
ENSMUST00000181562.8
|
Gm3488
|
predicted gene, 3488 |
| chr13_+_23930717 | 1.63 |
ENSMUST00000099703.5
|
H2bc3
|
H2B clustered histone 3 |
| chr3_+_134918298 | 1.53 |
ENSMUST00000062893.12
|
Cenpe
|
centromere protein E |
| chr3_-_19365431 | 1.51 |
ENSMUST00000099195.10
|
Pde7a
|
phosphodiesterase 7A |
| chrX_+_11178173 | 1.49 |
ENSMUST00000178979.2
|
H2al1e
|
H2A histone family member L1E |
| chr14_-_17742998 | 1.42 |
ENSMUST00000165619.8
|
Gm3252
|
predicted gene 3252 |
| chr13_-_103094784 | 1.42 |
ENSMUST00000172264.8
ENSMUST00000099202.10 |
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr13_+_52000704 | 1.41 |
ENSMUST00000021903.3
|
Gadd45g
|
growth arrest and DNA-damage-inducible 45 gamma |
| chr15_-_99717956 | 1.40 |
ENSMUST00000109024.9
|
Lima1
|
LIM domain and actin binding 1 |
| chr6_-_113354826 | 1.34 |
ENSMUST00000032410.14
|
Tada3
|
transcriptional adaptor 3 |
| chr6_-_36787096 | 1.33 |
ENSMUST00000201321.2
ENSMUST00000101534.5 |
Ptn
|
pleiotrophin |
| chr11_+_77353431 | 1.32 |
ENSMUST00000130255.2
|
Coro6
|
coronin 6 |
| chr7_+_119495058 | 1.25 |
ENSMUST00000106518.9
ENSMUST00000207270.2 ENSMUST00000208424.2 ENSMUST00000208202.2 ENSMUST00000054440.11 |
Lyrm1
|
LYR motif containing 1 |
| chr19_-_5962798 | 1.18 |
ENSMUST00000118623.2
|
Dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr14_+_15295240 | 1.16 |
ENSMUST00000172431.8
|
Gm3512
|
predicted gene 3512 |
| chr14_-_17398733 | 1.16 |
ENSMUST00000163719.8
|
Gm8281
|
predicted gene, 8281 |
| chr14_+_16361108 | 1.07 |
ENSMUST00000165193.2
|
Gm3468
|
predicted gene 3468 |
| chr2_-_152256947 | 1.02 |
ENSMUST00000099207.5
|
Zcchc3
|
zinc finger, CCHC domain containing 3 |
| chr7_-_44646645 | 1.02 |
ENSMUST00000207342.2
|
Bcl2l12
|
BCL2-like 12 (proline rich) |
| chr6_-_113354668 | 0.99 |
ENSMUST00000193384.2
|
Tada3
|
transcriptional adaptor 3 |
| chr1_-_44118902 | 0.96 |
ENSMUST00000238662.2
|
Gm8251
|
predicted gene 8251 |
| chr11_-_12362136 | 0.94 |
ENSMUST00000174874.8
|
Cobl
|
cordon-bleu WH2 repeat |
| chr7_+_119495515 | 0.93 |
ENSMUST00000106517.9
|
Lyrm1
|
LYR motif containing 1 |
| chr2_-_111100733 | 0.93 |
ENSMUST00000099619.6
|
Olfr1277
|
olfactory receptor 1277 |
| chr14_-_17614197 | 0.88 |
ENSMUST00000166776.8
|
Gm3264
|
predicted gene 3264 |
| chr19_-_5962862 | 0.87 |
ENSMUST00000136983.8
|
Dpf2
|
D4, zinc and double PHD fingers family 2 |
| chrX_+_11187731 | 0.85 |
ENSMUST00000177926.3
|
H2al1h
|
H2A histone family member L1H |
| chr14_-_18817743 | 0.82 |
ENSMUST00000167430.8
|
Gm3020
|
predicted gene 3020 |
| chr5_+_103902020 | 0.82 |
ENSMUST00000054979.10
|
Aff1
|
AF4/FMR2 family, member 1 |
| chr17_+_28259749 | 0.80 |
ENSMUST00000233869.2
|
Anks1
|
ankyrin repeat and SAM domain containing 1 |
| chr14_-_19420488 | 0.78 |
ENSMUST00000166494.2
|
Gm2897
|
predicted gene 2897 |
| chr14_-_19137146 | 0.76 |
ENSMUST00000177786.8
|
Gm2956
|
predicted gene 2956 |
| chr17_+_37756371 | 0.74 |
ENSMUST00000078207.4
ENSMUST00000218675.2 |
Olfr108
|
olfactory receptor 108 |
| chrX_+_11175063 | 0.73 |
ENSMUST00000178595.2
|
H2al1d
|
H2A histone family member L1D |
| chr15_-_77129706 | 0.69 |
ENSMUST00000228361.2
|
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr14_-_18359247 | 0.63 |
ENSMUST00000170207.8
|
Gm8108
|
predicted gene 8108 |
| chrY_-_10147183 | 0.63 |
ENSMUST00000189630.2
|
Gm21310
|
predicted gene, 21310 |
| chr14_-_19057159 | 0.62 |
ENSMUST00000170123.2
|
Gm10409
|
predicted gene 10409 |
| chr14_+_16728196 | 0.58 |
ENSMUST00000177556.8
|
Gm3373
|
predicted gene 3373 |
| chr14_+_15579811 | 0.53 |
ENSMUST00000171906.2
|
Gm3667
|
predicted gene 3667 |
| chr14_+_15154724 | 0.53 |
ENSMUST00000165744.2
|
Gm3739
|
predicted gene 3739 |
| chr14_-_19635203 | 0.53 |
ENSMUST00000170694.9
|
Gm2237
|
predicted gene 2237 |
| chr7_+_17743802 | 0.52 |
ENSMUST00000081703.8
ENSMUST00000108488.7 |
Ceacam13
|
carcinoembryonic antigen-related cell adhesion molecule 13 |
| chr9_+_75213570 | 0.51 |
ENSMUST00000213990.2
|
Gnb5
|
guanine nucleotide binding protein (G protein), beta 5 |
| chr9_+_72952115 | 0.51 |
ENSMUST00000184146.8
ENSMUST00000034722.5 |
Rab27a
|
RAB27A, member RAS oncogene family |
| chr13_+_109397184 | 0.39 |
ENSMUST00000153234.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr7_-_119494918 | 0.37 |
ENSMUST00000059851.14
|
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr14_-_18287197 | 0.36 |
ENSMUST00000164512.8
|
Gm2974
|
predicted gene 2974 |
| chr7_-_101859308 | 0.36 |
ENSMUST00000070165.7
ENSMUST00000211235.2 ENSMUST00000211022.2 |
Nup98
|
nucleoporin 98 |
| chr5_+_24618380 | 0.35 |
ENSMUST00000049346.10
|
Asic3
|
acid-sensing (proton-gated) ion channel 3 |
| chr16_-_4698148 | 0.35 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1 |
| chr7_-_119494669 | 0.34 |
ENSMUST00000098080.9
|
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr3_-_93846078 | 0.34 |
ENSMUST00000177735.2
|
Tdpoz9-ps1
|
TD and POZ domain containing 9, pseudogene 1 |
| chr14_+_16182428 | 0.28 |
ENSMUST00000170104.3
|
Gm3411
|
predicted gene 3411 |
| chr5_-_148336574 | 0.27 |
ENSMUST00000202457.4
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr6_-_97156032 | 0.25 |
ENSMUST00000095664.6
|
Tmf1
|
TATA element modulatory factor 1 |
| chr4_-_116228921 | 0.24 |
ENSMUST00000239239.2
ENSMUST00000239177.2 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr7_-_101859379 | 0.18 |
ENSMUST00000210682.2
|
Nup98
|
nucleoporin 98 |
| chr14_+_15442324 | 0.18 |
ENSMUST00000170738.3
|
Gm10406
|
predicted gene 10406 |
| chr7_-_101859033 | 0.17 |
ENSMUST00000211005.2
|
Nup98
|
nucleoporin 98 |
| chr14_+_17080724 | 0.17 |
ENSMUST00000177986.8
|
Gm3500
|
predicted gene 3500 |
| chr17_-_37508902 | 0.15 |
ENSMUST00000055324.8
|
Olfr94
|
olfactory receptor 94 |
| chr15_-_77129786 | 0.15 |
ENSMUST00000228558.2
|
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr19_+_12655487 | 0.13 |
ENSMUST00000215134.2
ENSMUST00000049724.8 |
Olfr1443
|
olfactory receptor 1443 |
| chr13_-_21823691 | 0.12 |
ENSMUST00000043081.3
|
Olfr11
|
olfactory receptor 11 |
| chr4_-_43025792 | 0.05 |
ENSMUST00000067481.6
|
Pigo
|
phosphatidylinositol glycan anchor biosynthesis, class O |
| chr8_+_95113066 | 0.02 |
ENSMUST00000161576.8
ENSMUST00000034220.8 |
Herpud1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr14_-_18897750 | 0.02 |
ENSMUST00000178728.2
|
Gm3005
|
predicted gene 3005 |
| chr4_-_43025756 | 0.01 |
ENSMUST00000098109.9
|
Pigo
|
phosphatidylinositol glycan anchor biosynthesis, class O |
| chr2_+_143757193 | 0.01 |
ENSMUST00000103172.4
|
Dstn
|
destrin |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.3 | 53.1 | GO:0007522 | visceral muscle development(GO:0007522) |
| 7.5 | 30.2 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 4.9 | 53.8 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 4.6 | 18.4 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 4.1 | 20.7 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 3.7 | 14.6 | GO:0030091 | protein repair(GO:0030091) |
| 3.2 | 19.0 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 2.9 | 8.7 | GO:0021998 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) positive regulation of cardiac ventricle development(GO:1904414) |
| 2.6 | 15.7 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 2.0 | 8.0 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 2.0 | 5.9 | GO:1903412 | response to bile acid(GO:1903412) cellular response to bile acid(GO:1903413) |
| 1.6 | 14.7 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 1.6 | 17.7 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 1.5 | 9.2 | GO:0006116 | NADH oxidation(GO:0006116) |
| 1.4 | 4.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 1.3 | 13.4 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 1.1 | 9.9 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 1.0 | 7.6 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.9 | 10.7 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.9 | 17.6 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.9 | 11.4 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.8 | 55.3 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.7 | 2.9 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.7 | 12.0 | GO:0033622 | integrin activation(GO:0033622) |
| 0.7 | 6.1 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.6 | 26.9 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.5 | 3.2 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.5 | 2.8 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.4 | 3.1 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.4 | 5.2 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.4 | 2.0 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.4 | 3.0 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.4 | 4.3 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.3 | 4.0 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.3 | 1.3 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.3 | 6.9 | GO:0000272 | polysaccharide catabolic process(GO:0000272) |
| 0.3 | 2.9 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.3 | 4.2 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.3 | 11.5 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.3 | 25.7 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 0.3 | 2.4 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.2 | 19.3 | GO:0042398 | cellular modified amino acid biosynthetic process(GO:0042398) |
| 0.2 | 6.3 | GO:0001553 | luteinization(GO:0001553) |
| 0.2 | 3.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 4.4 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.2 | 4.3 | GO:0000054 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.2 | 3.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 9.2 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.2 | 0.8 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.2 | 1.5 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.2 | 11.3 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.2 | 0.5 | GO:1903433 | regulation of constitutive secretory pathway(GO:1903433) |
| 0.2 | 3.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.2 | 0.9 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 3.5 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.1 | 4.6 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 13.9 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 0.7 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 12.8 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.1 | 9.8 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.1 | 3.0 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 1.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 1.1 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 1.8 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 2.3 | GO:0007614 | short-term memory(GO:0007614) |
| 0.1 | 0.8 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.2 | GO:2000845 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.1 | 0.4 | GO:0050915 | sensory perception of sour taste(GO:0050915) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.1 | 6.4 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.4 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.5 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 4.7 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.2 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 1.4 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.6 | GO:0051443 | protein neddylation(GO:0045116) positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 3.1 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.3 | GO:0015809 | arginine transport(GO:0015809) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.0 | 33.1 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 4.7 | 19.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 3.7 | 25.7 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 2.6 | 70.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 2.0 | 9.9 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 1.7 | 18.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 1.6 | 8.0 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 1.1 | 74.5 | GO:0031672 | A band(GO:0031672) |
| 0.9 | 15.7 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.9 | 13.9 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.8 | 4.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.7 | 11.4 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.5 | 11.4 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.5 | 2.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.5 | 57.4 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.5 | 14.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.4 | 7.3 | GO:0043034 | costamere(GO:0043034) |
| 0.4 | 63.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.3 | 3.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) RISC-loading complex(GO:0070578) |
| 0.3 | 2.9 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.3 | 3.4 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.3 | 20.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.2 | 2.0 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 0.9 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 13.7 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 0.7 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 28.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 10.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 9.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 6.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 1.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 8.7 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 7.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 6.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 24.1 | GO:0005925 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.0 | 2.1 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 5.4 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 4.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 11.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 9.6 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 2.2 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 6.9 | GO:0030424 | axon(GO:0030424) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.6 | 53.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 3.1 | 9.2 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 2.8 | 19.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 2.6 | 15.7 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 2.4 | 14.6 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 2.2 | 13.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 2.1 | 63.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 1.7 | 6.9 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 1.6 | 14.7 | GO:0004568 | chitinase activity(GO:0004568) |
| 1.3 | 8.0 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 1.3 | 9.2 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 1.0 | 15.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.9 | 8.7 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.8 | 5.0 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.8 | 17.6 | GO:0031005 | filamin binding(GO:0031005) |
| 0.7 | 7.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.7 | 29.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.7 | 2.0 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.6 | 18.4 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.5 | 4.6 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.4 | 9.4 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.4 | 20.7 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.4 | 33.6 | GO:0017022 | myosin binding(GO:0017022) |
| 0.4 | 3.2 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.4 | 2.4 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.4 | 2.9 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.3 | 3.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.3 | 1.3 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.3 | 10.7 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.3 | 4.1 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.3 | 4.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.3 | 3.4 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.3 | 2.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 12.8 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.2 | 1.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 35.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 6.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 11.9 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.2 | 12.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 4.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 4.0 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 3.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 1.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 25.7 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 4.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 4.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 68.0 | GO:0005524 | ATP binding(GO:0005524) |
| 0.0 | 0.5 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 1.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.3 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.4 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 8.4 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 4.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 4.3 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 38.2 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 11.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 4.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 2.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 67.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.8 | 30.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.6 | 33.3 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.5 | 15.7 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.4 | 9.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 19.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.2 | 5.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.2 | 10.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 8.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 8.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 4.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 10.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 6.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 3.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 1.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 2.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 4.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 158.4 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 1.0 | 29.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.6 | 18.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.5 | 15.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.5 | 10.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.4 | 6.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.3 | 8.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.2 | 4.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 32.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.2 | 5.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 39.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 8.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 3.2 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 3.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 19.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 9.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 3.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 2.4 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |