Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Stat1
Z-value: 0.80
Transcription factors associated with Stat1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Stat1
|
ENSMUSG00000026104.15 | Stat1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Stat1 | mm39_v1_chr1_+_52158721_52158783 | 0.07 | 5.4e-01 | Click! |
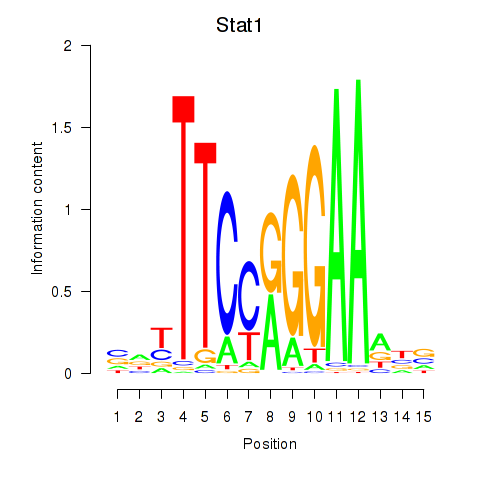
Activity profile of Stat1 motif
Sorted Z-values of Stat1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Stat1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_55950939 | 7.34 |
ENSMUST00000168729.8
ENSMUST00000228123.2 ENSMUST00000178034.9 |
Tgm1
|
transglutaminase 1, K polypeptide |
| chr5_+_14075281 | 6.30 |
ENSMUST00000073957.8
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr15_-_54783357 | 5.23 |
ENSMUST00000167541.3
ENSMUST00000171545.9 ENSMUST00000041591.16 ENSMUST00000173516.8 |
Enpp2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chrX_-_60229164 | 5.07 |
ENSMUST00000166381.3
|
Cdr1
|
cerebellar degeneration related antigen 1 |
| chr16_-_42160957 | 3.76 |
ENSMUST00000102817.5
|
Gap43
|
growth associated protein 43 |
| chr7_+_91321500 | 3.73 |
ENSMUST00000238619.2
ENSMUST00000238467.2 |
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chrX_+_7594670 | 3.51 |
ENSMUST00000033489.8
|
Praf2
|
PRA1 domain family 2 |
| chr7_+_43430459 | 3.46 |
ENSMUST00000014058.11
|
Klk10
|
kallikrein related-peptidase 10 |
| chr14_-_70864448 | 3.35 |
ENSMUST00000110984.4
|
Dmtn
|
dematin actin binding protein |
| chr7_+_91321694 | 3.32 |
ENSMUST00000238608.2
|
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr7_+_29607917 | 3.22 |
ENSMUST00000186475.2
|
Zfp383
|
zinc finger protein 383 |
| chr8_-_71345391 | 3.15 |
ENSMUST00000000809.3
|
Slc5a5
|
solute carrier family 5 (sodium iodide symporter), member 5 |
| chr3_+_14011445 | 3.06 |
ENSMUST00000192209.6
ENSMUST00000171075.8 ENSMUST00000108372.4 |
Ralyl
|
RALY RNA binding protein-like |
| chr15_-_37459570 | 3.04 |
ENSMUST00000119730.8
ENSMUST00000120746.8 |
Ncald
|
neurocalcin delta |
| chr9_+_59614877 | 3.03 |
ENSMUST00000128944.8
ENSMUST00000098661.10 |
Gramd2
|
GRAM domain containing 2 |
| chr14_-_70864666 | 3.02 |
ENSMUST00000022694.17
|
Dmtn
|
dematin actin binding protein |
| chr9_+_110867807 | 2.82 |
ENSMUST00000197575.2
|
Ltf
|
lactotransferrin |
| chr1_+_134110142 | 2.75 |
ENSMUST00000082060.10
ENSMUST00000153856.8 ENSMUST00000133701.8 ENSMUST00000132873.8 |
Chil1
|
chitinase-like 1 |
| chr9_+_45029080 | 2.69 |
ENSMUST00000170998.9
ENSMUST00000093855.4 |
Scn2b
|
sodium channel, voltage-gated, type II, beta |
| chr7_+_45433103 | 2.68 |
ENSMUST00000209617.2
ENSMUST00000209701.2 |
Lmtk3
|
lemur tyrosine kinase 3 |
| chr2_-_117173428 | 2.65 |
ENSMUST00000102534.11
|
Rasgrp1
|
RAS guanyl releasing protein 1 |
| chr2_-_117173312 | 2.60 |
ENSMUST00000178884.8
|
Rasgrp1
|
RAS guanyl releasing protein 1 |
| chr11_+_101136821 | 2.57 |
ENSMUST00000129680.8
|
Ramp2
|
receptor (calcitonin) activity modifying protein 2 |
| chr2_-_117173190 | 2.55 |
ENSMUST00000173541.8
ENSMUST00000172901.8 ENSMUST00000173252.2 |
Rasgrp1
|
RAS guanyl releasing protein 1 |
| chr14_+_84680993 | 2.51 |
ENSMUST00000071370.7
|
Pcdh17
|
protocadherin 17 |
| chr1_+_134109888 | 2.41 |
ENSMUST00000156873.8
|
Chil1
|
chitinase-like 1 |
| chr7_+_45204317 | 2.40 |
ENSMUST00000107752.12
|
Hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr15_-_37458768 | 2.39 |
ENSMUST00000116445.9
|
Ncald
|
neurocalcin delta |
| chr15_-_93417380 | 2.34 |
ENSMUST00000109255.3
|
Prickle1
|
prickle planar cell polarity protein 1 |
| chr7_+_45433306 | 2.29 |
ENSMUST00000072580.12
|
Lmtk3
|
lemur tyrosine kinase 3 |
| chr16_-_20245071 | 2.04 |
ENSMUST00000115547.9
ENSMUST00000096199.5 |
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr18_+_32087883 | 2.03 |
ENSMUST00000223753.2
|
Lims2
|
LIM and senescent cell antigen like domains 2 |
| chr7_+_107166653 | 2.02 |
ENSMUST00000120990.2
|
Olfml1
|
olfactomedin-like 1 |
| chr11_+_78389913 | 2.02 |
ENSMUST00000017488.5
|
Vtn
|
vitronectin |
| chr12_-_100486950 | 2.01 |
ENSMUST00000223020.2
ENSMUST00000062957.8 |
Ttc7b
|
tetratricopeptide repeat domain 7B |
| chr7_+_23954167 | 1.93 |
ENSMUST00000206777.2
|
Zfp108
|
zinc finger protein 108 |
| chr16_+_17327076 | 1.91 |
ENSMUST00000232242.2
|
Lztr1
|
leucine-zipper-like transcriptional regulator, 1 |
| chr2_+_177834868 | 1.89 |
ENSMUST00000103065.2
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr18_+_37453427 | 1.75 |
ENSMUST00000078271.4
|
Pcdhb5
|
protocadherin beta 5 |
| chr8_+_26022141 | 1.72 |
ENSMUST00000210846.2
ENSMUST00000167764.2 |
Fgfr1
|
fibroblast growth factor receptor 1 |
| chr7_+_107166925 | 1.69 |
ENSMUST00000239087.2
|
Olfml1
|
olfactomedin-like 1 |
| chr16_-_20244631 | 1.69 |
ENSMUST00000077867.10
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr16_-_20245138 | 1.68 |
ENSMUST00000079158.13
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr4_+_114916703 | 1.68 |
ENSMUST00000162489.2
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr8_-_71060911 | 1.67 |
ENSMUST00000210580.2
ENSMUST00000211608.2 ENSMUST00000049908.11 |
Ssbp4
|
single stranded DNA binding protein 4 |
| chr7_-_24016020 | 1.62 |
ENSMUST00000108436.8
ENSMUST00000032673.15 |
Zfp94
|
zinc finger protein 94 |
| chr9_+_123822000 | 1.60 |
ENSMUST00000039171.9
|
Ccr3
|
chemokine (C-C motif) receptor 3 |
| chr7_+_45204350 | 1.58 |
ENSMUST00000210300.2
|
Hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chrX_-_72974357 | 1.57 |
ENSMUST00000155597.2
ENSMUST00000114379.8 |
Renbp
|
renin binding protein |
| chrX_+_105059305 | 1.57 |
ENSMUST00000033582.5
|
Cox7b
|
cytochrome c oxidase subunit 7B |
| chr3_+_89136353 | 1.53 |
ENSMUST00000041142.4
|
Muc1
|
mucin 1, transmembrane |
| chr12_-_108859123 | 1.50 |
ENSMUST00000161154.2
ENSMUST00000161410.8 |
Wars
|
tryptophanyl-tRNA synthetase |
| chr16_+_17326810 | 1.49 |
ENSMUST00000231292.2
|
Lztr1
|
leucine-zipper-like transcriptional regulator, 1 |
| chr9_-_96601574 | 1.48 |
ENSMUST00000128269.8
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chr3_-_57202546 | 1.46 |
ENSMUST00000196506.2
|
Tm4sf1
|
transmembrane 4 superfamily member 1 |
| chr6_-_65121892 | 1.46 |
ENSMUST00000031982.5
|
Hpgds
|
hematopoietic prostaglandin D synthase |
| chr3_-_57202301 | 1.45 |
ENSMUST00000171384.8
|
Tm4sf1
|
transmembrane 4 superfamily member 1 |
| chr8_+_105996469 | 1.44 |
ENSMUST00000172525.8
ENSMUST00000174837.8 ENSMUST00000173859.2 |
Hsf4
|
heat shock transcription factor 4 |
| chr19_-_46315543 | 1.41 |
ENSMUST00000223917.2
ENSMUST00000224447.2 ENSMUST00000041391.5 ENSMUST00000096029.12 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr9_-_71393175 | 1.41 |
ENSMUST00000233263.2
ENSMUST00000034720.12 |
Polr2m
|
polymerase (RNA) II (DNA directed) polypeptide M |
| chr7_+_29467971 | 1.37 |
ENSMUST00000032802.5
|
Zfp84
|
zinc finger protein 84 |
| chr3_-_10505113 | 1.37 |
ENSMUST00000029047.12
ENSMUST00000195822.2 ENSMUST00000099223.11 |
Snx16
|
sorting nexin 16 |
| chr3_-_96201248 | 1.36 |
ENSMUST00000029748.8
|
Fcgr1
|
Fc receptor, IgG, high affinity I |
| chr10_+_69761784 | 1.36 |
ENSMUST00000181974.8
ENSMUST00000182795.8 ENSMUST00000182437.8 |
Ank3
|
ankyrin 3, epithelial |
| chrX_-_8011952 | 1.31 |
ENSMUST00000115615.9
ENSMUST00000115616.8 ENSMUST00000115621.9 |
Rbm3
|
RNA binding motif (RNP1, RRM) protein 3 |
| chrX_-_8011918 | 1.30 |
ENSMUST00000115619.8
ENSMUST00000115617.10 ENSMUST00000040010.10 |
Rbm3
|
RNA binding motif (RNP1, RRM) protein 3 |
| chr10_+_69761597 | 1.30 |
ENSMUST00000182269.8
ENSMUST00000183261.8 ENSMUST00000183074.8 |
Ank3
|
ankyrin 3, epithelial |
| chr12_-_112637998 | 1.30 |
ENSMUST00000128300.9
|
Akt1
|
thymoma viral proto-oncogene 1 |
| chr1_-_37929414 | 1.29 |
ENSMUST00000139725.8
ENSMUST00000027257.10 |
Mitd1
|
MIT, microtubule interacting and transport, domain containing 1 |
| chr3_+_84832783 | 1.27 |
ENSMUST00000107675.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr4_-_129472328 | 1.26 |
ENSMUST00000052835.9
|
Fam167b
|
family with sequence similarity 167, member B |
| chr11_+_3438274 | 1.26 |
ENSMUST00000064265.13
|
Pla2g3
|
phospholipase A2, group III |
| chr9_+_32135781 | 1.25 |
ENSMUST00000183121.2
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chrX_-_72974440 | 1.24 |
ENSMUST00000116578.8
|
Renbp
|
renin binding protein |
| chr4_+_120523758 | 1.24 |
ENSMUST00000094814.6
|
Cited4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chr9_-_75466865 | 1.24 |
ENSMUST00000215821.2
|
Tmod3
|
tropomodulin 3 |
| chr7_-_23907518 | 1.22 |
ENSMUST00000086006.12
|
Zfp111
|
zinc finger protein 111 |
| chr8_-_71229293 | 1.20 |
ENSMUST00000034296.15
|
Pik3r2
|
phosphoinositide-3-kinase regulatory subunit 2 |
| chr7_+_23833572 | 1.20 |
ENSMUST00000205680.2
ENSMUST00000056549.9 |
Zfp235
|
zinc finger protein 235 |
| chr7_-_126224848 | 1.19 |
ENSMUST00000032961.4
|
Nupr1
|
nuclear protein transcription regulator 1 |
| chr11_+_81992662 | 1.17 |
ENSMUST00000000194.4
|
Ccl12
|
chemokine (C-C motif) ligand 12 |
| chr9_+_105520154 | 1.15 |
ENSMUST00000190358.2
ENSMUST00000191268.7 ENSMUST00000065778.13 ENSMUST00000188784.2 |
Pik3r4
|
phosphoinositide-3-kinase regulatory subunit 4 |
| chr5_+_67418137 | 1.12 |
ENSMUST00000161369.3
|
Tmem33
|
transmembrane protein 33 |
| chr16_+_19578981 | 1.09 |
ENSMUST00000079780.10
ENSMUST00000164397.8 |
B3gnt5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr10_-_61619790 | 1.08 |
ENSMUST00000020283.5
|
Macroh2a2
|
macroH2A.2 histone |
| chr13_+_83720484 | 1.06 |
ENSMUST00000196207.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr7_+_29515485 | 1.06 |
ENSMUST00000178162.3
ENSMUST00000032796.14 |
Zfp790
|
zinc finger protein 790 |
| chr5_+_102629365 | 1.04 |
ENSMUST00000112854.8
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr3_+_142202642 | 1.04 |
ENSMUST00000090127.7
|
Gbp5
|
guanylate binding protein 5 |
| chr7_-_100505486 | 1.03 |
ENSMUST00000139604.8
|
Relt
|
RELT tumor necrosis factor receptor |
| chr17_+_36134398 | 1.01 |
ENSMUST00000173493.8
ENSMUST00000173147.8 |
Flot1
|
flotillin 1 |
| chr16_+_91169770 | 1.01 |
ENSMUST00000089042.7
|
Ifnar2
|
interferon (alpha and beta) receptor 2 |
| chr7_+_29794575 | 1.00 |
ENSMUST00000130526.2
ENSMUST00000108200.2 |
Zfp260
|
zinc finger protein 260 |
| chr7_+_23811739 | 0.98 |
ENSMUST00000120006.8
ENSMUST00000005413.4 |
Zfp112
|
zinc finger protein 112 |
| chr13_+_83720457 | 0.97 |
ENSMUST00000196730.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr10_-_78427721 | 0.95 |
ENSMUST00000040580.7
|
Syde1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans) |
| chr11_+_101315893 | 0.93 |
ENSMUST00000040561.6
|
Rundc1
|
RUN domain containing 1 |
| chr10_+_69234810 | 0.93 |
ENSMUST00000218680.2
|
Ank3
|
ankyrin 3, epithelial |
| chr14_-_45767232 | 0.92 |
ENSMUST00000149723.2
|
Fermt2
|
fermitin family member 2 |
| chr16_+_19578945 | 0.92 |
ENSMUST00000121344.8
|
B3gnt5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chrX_-_46981273 | 0.89 |
ENSMUST00000153548.9
ENSMUST00000141084.3 |
Smarca1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr17_+_36134450 | 0.88 |
ENSMUST00000172846.2
|
Flot1
|
flotillin 1 |
| chr7_-_12819142 | 0.83 |
ENSMUST00000094829.2
|
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr9_+_45230370 | 0.83 |
ENSMUST00000034597.8
|
Tmprss13
|
transmembrane protease, serine 13 |
| chr4_-_136613498 | 0.83 |
ENSMUST00000046384.9
|
C1qb
|
complement component 1, q subcomponent, beta polypeptide |
| chr11_-_98620200 | 0.82 |
ENSMUST00000126565.2
ENSMUST00000100500.9 ENSMUST00000017354.13 |
Med24
|
mediator complex subunit 24 |
| chr4_-_140344373 | 0.81 |
ENSMUST00000154979.2
|
Arhgef10l
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr4_+_114945905 | 0.80 |
ENSMUST00000171877.8
ENSMUST00000177647.8 ENSMUST00000106548.9 ENSMUST00000030488.3 |
Pdzk1ip1
|
PDZK1 interacting protein 1 |
| chr6_+_34575435 | 0.80 |
ENSMUST00000079391.10
ENSMUST00000142512.8 ENSMUST00000115027.8 ENSMUST00000115026.8 |
Cald1
|
caldesmon 1 |
| chr18_-_33596468 | 0.78 |
ENSMUST00000171533.9
|
Nrep
|
neuronal regeneration related protein |
| chr9_-_44437694 | 0.78 |
ENSMUST00000062215.8
|
Cxcr5
|
chemokine (C-X-C motif) receptor 5 |
| chr9_+_38629560 | 0.78 |
ENSMUST00000001544.12
ENSMUST00000118144.8 |
Vwa5a
|
von Willebrand factor A domain containing 5A |
| chr5_-_34671236 | 0.78 |
ENSMUST00000114359.2
ENSMUST00000030991.14 ENSMUST00000087737.10 |
Tnip2
|
TNFAIP3 interacting protein 2 |
| chr1_-_54596754 | 0.78 |
ENSMUST00000097739.5
|
Pgap1
|
post-GPI attachment to proteins 1 |
| chr7_-_44665639 | 0.77 |
ENSMUST00000085383.11
|
Scaf1
|
SR-related CTD-associated factor 1 |
| chr3_-_88417251 | 0.76 |
ENSMUST00000149068.2
|
Lmna
|
lamin A |
| chr15_-_97729341 | 0.75 |
ENSMUST00000079838.14
ENSMUST00000118294.8 |
Hdac7
|
histone deacetylase 7 |
| chr10_+_40225272 | 0.75 |
ENSMUST00000044672.11
ENSMUST00000095743.4 |
Cdk19
|
cyclin-dependent kinase 19 |
| chr9_-_117701613 | 0.75 |
ENSMUST00000239475.2
|
Rbms3
|
RNA binding motif, single stranded interacting protein |
| chr17_+_34364206 | 0.74 |
ENSMUST00000041982.9
ENSMUST00000171231.8 |
H2-DMb2
|
histocompatibility 2, class II, locus Mb2 |
| chr16_+_91169671 | 0.74 |
ENSMUST00000023693.14
ENSMUST00000134491.9 ENSMUST00000117836.8 |
Ifnar2
|
interferon (alpha and beta) receptor 2 |
| chr7_-_24245419 | 0.73 |
ENSMUST00000011776.8
|
Pinlyp
|
phospholipase A2 inhibitor and LY6/PLAUR domain containing |
| chr17_+_28910393 | 0.73 |
ENSMUST00000124886.9
ENSMUST00000114758.9 |
Mapk14
|
mitogen-activated protein kinase 14 |
| chrX_-_8118541 | 0.72 |
ENSMUST00000115594.8
ENSMUST00000115595.8 ENSMUST00000033513.10 |
Ftsj1
|
FtsJ RNA methyltransferase homolog 1 (E. coli) |
| chr6_+_41498716 | 0.71 |
ENSMUST00000070380.5
|
Prss2
|
protease, serine 2 |
| chr7_+_81984940 | 0.70 |
ENSMUST00000173287.8
|
Adamtsl3
|
ADAMTS-like 3 |
| chr7_-_108774367 | 0.69 |
ENSMUST00000207178.2
|
Lmo1
|
LIM domain only 1 |
| chr7_+_23969822 | 0.69 |
ENSMUST00000108438.10
|
Zfp93
|
zinc finger protein 93 |
| chr4_+_148686985 | 0.69 |
ENSMUST00000105701.9
ENSMUST00000052060.7 |
Masp2
|
mannan-binding lectin serine peptidase 2 |
| chr17_+_28910302 | 0.68 |
ENSMUST00000004990.14
ENSMUST00000114754.8 ENSMUST00000062694.16 |
Mapk14
|
mitogen-activated protein kinase 14 |
| chr18_-_33596890 | 0.67 |
ENSMUST00000237066.2
|
Nrep
|
neuronal regeneration related protein |
| chr14_-_118943591 | 0.66 |
ENSMUST00000036554.14
ENSMUST00000166646.2 |
Abcc4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
| chr5_+_67417908 | 0.66 |
ENSMUST00000037918.12
ENSMUST00000162543.8 ENSMUST00000161233.8 ENSMUST00000160352.8 |
Tmem33
|
transmembrane protein 33 |
| chr2_+_3425159 | 0.63 |
ENSMUST00000100463.10
ENSMUST00000061852.12 ENSMUST00000102988.10 ENSMUST00000115066.8 |
Dclre1c
|
DNA cross-link repair 1C |
| chr8_-_68363564 | 0.63 |
ENSMUST00000093468.12
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr7_+_65343156 | 0.63 |
ENSMUST00000032726.14
ENSMUST00000107495.5 ENSMUST00000143508.3 ENSMUST00000129166.3 ENSMUST00000206517.2 ENSMUST00000206837.2 ENSMUST00000206628.2 ENSMUST00000206361.2 |
Tm2d3
|
TM2 domain containing 3 |
| chr10_+_69761630 | 0.63 |
ENSMUST00000182029.8
|
Ank3
|
ankyrin 3, epithelial |
| chr10_-_126906123 | 0.62 |
ENSMUST00000060991.6
|
Tspan31
|
tetraspanin 31 |
| chr2_+_101716577 | 0.62 |
ENSMUST00000028584.8
|
Commd9
|
COMM domain containing 9 |
| chr9_-_44437801 | 0.61 |
ENSMUST00000215661.2
|
Cxcr5
|
chemokine (C-X-C motif) receptor 5 |
| chrX_-_99670174 | 0.61 |
ENSMUST00000015812.12
|
Pdzd11
|
PDZ domain containing 11 |
| chr2_-_120801186 | 0.59 |
ENSMUST00000028728.6
|
Ubr1
|
ubiquitin protein ligase E3 component n-recognin 1 |
| chr18_-_52662728 | 0.57 |
ENSMUST00000025409.9
|
Lox
|
lysyl oxidase |
| chr17_+_36134122 | 0.55 |
ENSMUST00000001569.15
ENSMUST00000174080.8 |
Flot1
|
flotillin 1 |
| chr14_+_55842002 | 0.55 |
ENSMUST00000138037.2
|
Irf9
|
interferon regulatory factor 9 |
| chr1_+_37929558 | 0.54 |
ENSMUST00000027256.12
ENSMUST00000193673.6 ENSMUST00000160082.3 |
Mrpl30
|
mitochondrial ribosomal protein L30 |
| chr2_+_128942919 | 0.53 |
ENSMUST00000028874.8
|
Polr1b
|
polymerase (RNA) I polypeptide B |
| chr2_-_127426588 | 0.52 |
ENSMUST00000110368.9
ENSMUST00000077422.12 |
Zfp661
|
zinc finger protein 661 |
| chr1_+_34044940 | 0.52 |
ENSMUST00000187486.7
ENSMUST00000182697.8 |
Dst
|
dystonin |
| chr9_+_107174081 | 0.51 |
ENSMUST00000167072.2
|
Cish
|
cytokine inducible SH2-containing protein |
| chr5_+_75312939 | 0.51 |
ENSMUST00000202681.4
ENSMUST00000000476.15 |
Pdgfra
|
platelet derived growth factor receptor, alpha polypeptide |
| chr3_+_96484294 | 0.49 |
ENSMUST00000148290.2
|
Gm16253
|
predicted gene 16253 |
| chrX_+_158086253 | 0.48 |
ENSMUST00000112491.2
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr16_-_59138611 | 0.44 |
ENSMUST00000216261.2
|
Olfr204
|
olfactory receptor 204 |
| chr2_+_128942900 | 0.44 |
ENSMUST00000103205.11
|
Polr1b
|
polymerase (RNA) I polypeptide B |
| chr9_-_30833748 | 0.43 |
ENSMUST00000065112.7
|
Adamts15
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 15 |
| chr16_+_21613068 | 0.42 |
ENSMUST00000211443.2
ENSMUST00000231300.2 ENSMUST00000209449.2 ENSMUST00000181780.9 ENSMUST00000209728.2 ENSMUST00000181960.3 ENSMUST00000209429.2 ENSMUST00000180830.3 ENSMUST00000231988.2 |
1300002E11Rik
Map3k13
|
RIKEN cDNA 1300002E11 gene mitogen-activated protein kinase kinase kinase 13 |
| chr19_-_46950355 | 0.42 |
ENSMUST00000236501.2
|
Nt5c2
|
5'-nucleotidase, cytosolic II |
| chr2_-_119916159 | 0.42 |
ENSMUST00000156159.4
|
Sptbn5
|
spectrin beta, non-erythrocytic 5 |
| chr5_+_89034666 | 0.41 |
ENSMUST00000148750.8
|
Slc4a4
|
solute carrier family 4 (anion exchanger), member 4 |
| chr11_-_75081284 | 0.41 |
ENSMUST00000044949.11
|
Dph1
|
diphthamide biosynthesis 1 |
| chr4_+_53826013 | 0.40 |
ENSMUST00000030127.13
|
Tmem38b
|
transmembrane protein 38B |
| chr12_-_103597663 | 0.40 |
ENSMUST00000121625.2
ENSMUST00000044231.12 |
Serpina10
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr17_-_48145466 | 0.38 |
ENSMUST00000066368.13
|
Mdfi
|
MyoD family inhibitor |
| chr7_-_100232276 | 0.38 |
ENSMUST00000152876.3
ENSMUST00000150042.8 ENSMUST00000132888.9 |
Mrpl48
|
mitochondrial ribosomal protein L48 |
| chr1_-_155293075 | 0.37 |
ENSMUST00000027741.12
|
Xpr1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr5_-_6926523 | 0.37 |
ENSMUST00000164784.2
|
Zfp804b
|
zinc finger protein 804B |
| chr6_-_122259768 | 0.37 |
ENSMUST00000032207.9
|
Klrg1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr4_-_129534752 | 0.36 |
ENSMUST00000132217.8
ENSMUST00000130017.2 ENSMUST00000154105.8 |
Txlna
|
taxilin alpha |
| chr13_-_67903312 | 0.35 |
ENSMUST00000144183.3
|
Zfp85
|
zinc finger protein 85 |
| chr8_-_111808674 | 0.34 |
ENSMUST00000039597.14
|
Clec18a
|
C-type lectin domain family 18, member A |
| chr9_-_123507937 | 0.34 |
ENSMUST00000040960.13
|
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr5_-_5564730 | 0.33 |
ENSMUST00000115445.8
ENSMUST00000179804.8 ENSMUST00000125110.2 ENSMUST00000115446.8 |
Cldn12
|
claudin 12 |
| chr5_-_148988110 | 0.33 |
ENSMUST00000110505.8
|
Hmgb1
|
high mobility group box 1 |
| chr11_-_45846291 | 0.32 |
ENSMUST00000011398.13
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr2_+_164245114 | 0.31 |
ENSMUST00000017151.2
|
Rbpjl
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr2_-_86640362 | 0.31 |
ENSMUST00000216117.2
|
Olfr141
|
olfactory receptor 141 |
| chr6_-_48025845 | 0.31 |
ENSMUST00000095944.10
|
Zfp777
|
zinc finger protein 777 |
| chr5_-_5564873 | 0.31 |
ENSMUST00000060947.14
|
Cldn12
|
claudin 12 |
| chr1_-_155293141 | 0.31 |
ENSMUST00000111775.8
ENSMUST00000111774.2 |
Xpr1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr5_+_134212836 | 0.30 |
ENSMUST00000016086.10
|
Gtf2ird2
|
GTF2I repeat domain containing 2 |
| chr19_-_37184692 | 0.29 |
ENSMUST00000132580.8
ENSMUST00000079754.11 ENSMUST00000136286.8 ENSMUST00000126188.8 ENSMUST00000126781.2 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr5_-_140687995 | 0.29 |
ENSMUST00000135028.5
ENSMUST00000077890.12 ENSMUST00000041783.14 ENSMUST00000142081.6 |
Iqce
|
IQ motif containing E |
| chr18_-_38417390 | 0.29 |
ENSMUST00000025311.8
|
Pcdh12
|
protocadherin 12 |
| chr3_-_90398270 | 0.26 |
ENSMUST00000149884.2
|
Snapin
|
SNAP-associated protein |
| chr17_+_29309942 | 0.26 |
ENSMUST00000119901.9
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chr4_-_107889267 | 0.24 |
ENSMUST00000106709.9
|
Podn
|
podocan |
| chr18_-_33596792 | 0.23 |
ENSMUST00000051087.16
|
Nrep
|
neuronal regeneration related protein |
| chr6_-_122317484 | 0.23 |
ENSMUST00000112600.9
|
Phc1
|
polyhomeotic 1 |
| chr7_+_6346723 | 0.23 |
ENSMUST00000207173.3
|
Gm3854
|
predicted gene 3854 |
| chr5_+_52991351 | 0.22 |
ENSMUST00000031072.14
|
Anapc4
|
anaphase promoting complex subunit 4 |
| chr13_+_110039620 | 0.21 |
ENSMUST00000120664.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr4_+_83335947 | 0.20 |
ENSMUST00000030206.10
ENSMUST00000071544.11 |
Snapc3
|
small nuclear RNA activating complex, polypeptide 3 |
| chr11_+_81926394 | 0.19 |
ENSMUST00000000193.6
|
Ccl2
|
chemokine (C-C motif) ligand 2 |
| chr6_+_40941688 | 0.18 |
ENSMUST00000076638.7
|
1810009J06Rik
|
RIKEN cDNA 1810009J06 gene |
| chr8_+_26091607 | 0.18 |
ENSMUST00000155861.8
|
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr6_+_129374441 | 0.17 |
ENSMUST00000112081.9
ENSMUST00000112079.3 |
Clec1b
|
C-type lectin domain family 1, member b |
| chr18_-_38417444 | 0.17 |
ENSMUST00000194012.2
|
Pcdh12
|
protocadherin 12 |
| chr18_-_52662917 | 0.15 |
ENSMUST00000171470.8
|
Lox
|
lysyl oxidase |
| chr1_+_173877941 | 0.15 |
ENSMUST00000062665.4
|
Olfr432
|
olfactory receptor 432 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 6.4 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.8 | 2.3 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.7 | 2.8 | GO:1900190 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.7 | 5.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.6 | 7.3 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.6 | 1.8 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.6 | 1.7 | GO:1903465 | motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.6 | 1.7 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.5 | 2.7 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.5 | 7.8 | GO:0032725 | positive regulation of granulocyte macrophage colony-stimulating factor production(GO:0032725) |
| 0.5 | 1.4 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.4 | 2.4 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.4 | 2.8 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.4 | 3.8 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.4 | 1.5 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.3 | 1.3 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.3 | 4.2 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.3 | 2.6 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.3 | 7.0 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.3 | 3.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.3 | 1.4 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.3 | 1.3 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.2 | 1.5 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.2 | 1.0 | GO:0017126 | nucleologenesis(GO:0017126) |
| 0.2 | 1.2 | GO:2000501 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.2 | 1.0 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.2 | 2.0 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.2 | 1.4 | GO:2001184 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.2 | 2.0 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.2 | 5.2 | GO:0072606 | interleukin-8 secretion(GO:0072606) |
| 0.2 | 2.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) liver regeneration(GO:0097421) |
| 0.1 | 0.6 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 2.5 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.1 | 6.3 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.1 | 4.0 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.1 | 1.8 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.1 | 0.8 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.5 | GO:0072277 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 1.0 | GO:0032621 | interleukin-18 production(GO:0032621) positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.1 | 0.3 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.1 | 1.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 1.3 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.1 | 1.0 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 0.6 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.3 | GO:1900248 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.1 | 0.3 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.1 | 0.4 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.6 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 0.8 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.1 | 0.8 | GO:0015791 | polyol transport(GO:0015791) |
| 0.1 | 1.4 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 1.2 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.1 | 1.7 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.1 | 1.4 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.7 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 2.0 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 2.6 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.1 | 2.0 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 0.3 | GO:1902774 | terminal button organization(GO:0072553) late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.7 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) phosphate ion transmembrane transport(GO:0035435) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.8 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 5.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.8 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.7 | GO:0032310 | prostaglandin secretion(GO:0032310) |
| 0.0 | 0.7 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.7 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.8 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.5 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 1.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 1.3 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 1.2 | GO:0042993 | positive regulation of transcription factor import into nucleus(GO:0042993) |
| 0.0 | 0.9 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.2 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 0.5 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.3 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.2 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.0 | 0.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.4 | GO:0046040 | IMP metabolic process(GO:0046040) |
| 0.0 | 0.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 2.2 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.3 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.4 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.9 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 1.4 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.0 | 0.1 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.8 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 6.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.6 | 1.7 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.5 | 2.8 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.4 | 2.6 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.3 | 1.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.3 | 0.9 | GO:0090537 | CERF complex(GO:0090537) |
| 0.3 | 2.0 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.2 | 1.2 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.2 | 7.0 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 4.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.2 | 1.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 0.7 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 2.4 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.8 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 3.8 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 2.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.3 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 0.8 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 1.6 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 1.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 1.0 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.5 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.2 | GO:0044299 | C-fiber(GO:0044299) |
| 0.1 | 0.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.6 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 1.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.4 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 2.0 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 2.6 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.0 | 1.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 20.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.2 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.7 | 2.0 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.7 | 7.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.7 | 7.8 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.6 | 4.0 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.5 | 3.2 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.5 | 3.8 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.4 | 5.2 | GO:0008061 | chitin binding(GO:0008061) |
| 0.4 | 2.6 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.4 | 1.5 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.4 | 1.5 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.3 | 1.7 | GO:0004905 | interferon receptor activity(GO:0004904) type I interferon receptor activity(GO:0004905) type I interferon binding(GO:0019962) |
| 0.3 | 1.4 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.3 | 2.7 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.3 | 6.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.3 | 2.6 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.3 | 7.0 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 1.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.2 | 1.4 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.2 | 0.7 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.2 | 0.5 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.2 | 2.8 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.2 | 1.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 11.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.6 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 1.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.7 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 1.4 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.3 | GO:0000401 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.1 | 5.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.6 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 1.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 2.8 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 0.3 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 0.9 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 0.6 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 2.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 2.0 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 1.0 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.7 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.1 | 1.3 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.1 | 0.7 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 2.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 2.4 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 1.6 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 2.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.4 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.7 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.7 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.8 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.7 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.7 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.3 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 1.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.0 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 1.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 5.8 | GO:0008514 | organic anion transmembrane transporter activity(GO:0008514) |
| 0.0 | 0.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.8 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 1.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.9 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 4.0 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 5.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 2.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 2.7 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 0.6 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.5 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 2.7 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 8.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.1 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.5 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 7.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.3 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.4 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.8 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.4 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.3 | 5.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 6.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 7.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 1.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 3.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 7.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.7 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 2.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.7 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 1.7 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 2.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 1.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 0.7 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 1.2 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.0 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.8 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 2.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |