Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
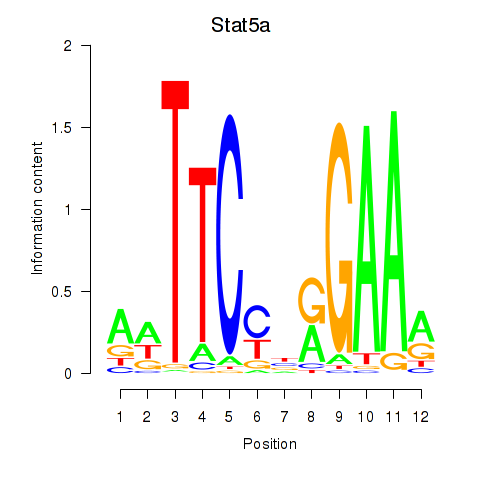
Results for Stat5a
Z-value: 2.28
Transcription factors associated with Stat5a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Stat5a
|
ENSMUSG00000004043.15 | Stat5a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Stat5a | mm39_v1_chr11_+_100750177_100750312 | 0.07 | 5.4e-01 | Click! |
Activity profile of Stat5a motif
Sorted Z-values of Stat5a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Stat5a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_87054796 | 16.98 |
ENSMUST00000031181.16
ENSMUST00000113333.2 |
Ugt2b34
|
UDP glucuronosyltransferase 2 family, polypeptide B34 |
| chr6_+_78347636 | 14.51 |
ENSMUST00000204873.3
|
Reg3b
|
regenerating islet-derived 3 beta |
| chr6_+_18170686 | 12.81 |
ENSMUST00000045706.12
|
Cftr
|
cystic fibrosis transmembrane conductance regulator |
| chr6_+_78347844 | 12.75 |
ENSMUST00000096904.6
ENSMUST00000203266.2 |
Reg3b
|
regenerating islet-derived 3 beta |
| chr7_-_28947882 | 12.68 |
ENSMUST00000032808.6
|
2200002D01Rik
|
RIKEN cDNA 2200002D01 gene |
| chr11_+_69855584 | 12.62 |
ENSMUST00000108597.8
ENSMUST00000060651.6 ENSMUST00000108596.8 |
Cldn7
|
claudin 7 |
| chr18_-_64688271 | 11.61 |
ENSMUST00000235459.2
|
Atp8b1
|
ATPase, class I, type 8B, member 1 |
| chr12_+_36042899 | 11.51 |
ENSMUST00000020898.12
|
Agr2
|
anterior gradient 2 |
| chr1_+_13738967 | 10.96 |
ENSMUST00000088542.4
|
Xkr9
|
X-linked Kx blood group related 9 |
| chr4_-_119515978 | 10.94 |
ENSMUST00000106309.9
ENSMUST00000044426.8 |
Guca2b
|
guanylate cyclase activator 2b (retina) |
| chr11_+_69856222 | 10.87 |
ENSMUST00000018713.13
|
Cldn7
|
claudin 7 |
| chr11_+_58269862 | 10.81 |
ENSMUST00000013787.11
ENSMUST00000108826.3 |
Lypd8
|
LY6/PLAUR domain containing 8 |
| chr6_-_83504756 | 10.47 |
ENSMUST00000152029.2
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr16_-_13548307 | 10.14 |
ENSMUST00000115807.9
|
Pla2g10
|
phospholipase A2, group X |
| chr11_+_98303287 | 10.06 |
ENSMUST00000058295.6
|
Erbb2
|
erb-b2 receptor tyrosine kinase 2 |
| chr3_+_10431961 | 9.93 |
ENSMUST00000029049.7
|
Chmp4c
|
charged multivesicular body protein 4C |
| chr10_+_115653152 | 9.80 |
ENSMUST00000080630.11
ENSMUST00000179196.3 ENSMUST00000035563.15 |
Tspan8
|
tetraspanin 8 |
| chr5_-_87682972 | 9.73 |
ENSMUST00000120150.2
|
Sult1b1
|
sulfotransferase family 1B, member 1 |
| chr6_+_18170781 | 9.59 |
ENSMUST00000115406.2
|
Cftr
|
cystic fibrosis transmembrane conductance regulator |
| chr1_+_88015524 | 9.56 |
ENSMUST00000113139.2
|
Ugt1a8
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr13_+_38335232 | 9.46 |
ENSMUST00000124830.3
|
Dsp
|
desmoplakin |
| chr13_-_32522548 | 9.44 |
ENSMUST00000041859.9
|
Gmds
|
GDP-mannose 4, 6-dehydratase |
| chr6_-_78445846 | 9.31 |
ENSMUST00000032089.3
|
Reg3g
|
regenerating islet-derived 3 gamma |
| chr16_-_13548833 | 9.16 |
ENSMUST00000023364.7
|
Pla2g10
|
phospholipase A2, group X |
| chr8_+_105652867 | 8.89 |
ENSMUST00000034355.11
ENSMUST00000109410.4 |
Ces2e
|
carboxylesterase 2E |
| chr4_-_55532453 | 8.82 |
ENSMUST00000132746.2
ENSMUST00000107619.3 |
Klf4
|
Kruppel-like factor 4 (gut) |
| chr13_-_4329421 | 8.76 |
ENSMUST00000021632.5
|
Akr1c12
|
aldo-keto reductase family 1, member C12 |
| chr6_-_86646118 | 8.70 |
ENSMUST00000001184.10
|
Mxd1
|
MAX dimerization protein 1 |
| chr6_-_83504471 | 8.65 |
ENSMUST00000141904.8
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr12_+_31488208 | 8.58 |
ENSMUST00000001254.6
|
Slc26a3
|
solute carrier family 26, member 3 |
| chr1_+_88022776 | 8.54 |
ENSMUST00000150634.8
ENSMUST00000058237.14 |
Ugt1a7c
|
UDP glucuronosyltransferase 1 family, polypeptide A7C |
| chr8_+_21777425 | 8.37 |
ENSMUST00000098893.4
|
Defa3
|
defensin, alpha, 3 |
| chr9_-_65330231 | 8.33 |
ENSMUST00000065894.7
|
Slc51b
|
solute carrier family 51, beta subunit |
| chr3_+_122688721 | 8.30 |
ENSMUST00000023820.6
|
Fabp2
|
fatty acid binding protein 2, intestinal |
| chr1_+_192835414 | 8.28 |
ENSMUST00000076521.7
|
Irf6
|
interferon regulatory factor 6 |
| chr4_-_154384464 | 8.02 |
ENSMUST00000030898.12
|
Arhgef16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr9_-_95697441 | 7.88 |
ENSMUST00000119760.2
|
Pls1
|
plastin 1 (I-isoform) |
| chr3_-_121325887 | 7.72 |
ENSMUST00000039197.9
|
Slc44a3
|
solute carrier family 44, member 3 |
| chr11_+_96822213 | 7.63 |
ENSMUST00000107633.2
|
Prr15l
|
proline rich 15-like |
| chr11_-_117859997 | 7.60 |
ENSMUST00000054002.4
|
Socs3
|
suppressor of cytokine signaling 3 |
| chr1_+_88128323 | 7.58 |
ENSMUST00000049289.9
|
Ugt1a2
|
UDP glucuronosyltransferase 1 family, polypeptide A2 |
| chr1_+_131566044 | 7.55 |
ENSMUST00000073350.13
|
Ctse
|
cathepsin E |
| chr11_-_120971954 | 7.52 |
ENSMUST00000106120.3
ENSMUST00000100126.9 ENSMUST00000106119.9 |
Sectm1a
|
secreted and transmembrane 1A |
| chr2_+_122478882 | 7.45 |
ENSMUST00000142767.8
|
AA467197
|
expressed sequence AA467197 |
| chr19_+_56276375 | 7.42 |
ENSMUST00000166049.8
|
Habp2
|
hyaluronic acid binding protein 2 |
| chr6_-_83513184 | 7.41 |
ENSMUST00000205926.2
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr3_+_69129745 | 7.40 |
ENSMUST00000183126.2
|
Arl14
|
ADP-ribosylation factor-like 14 |
| chr7_-_127529238 | 7.33 |
ENSMUST00000032988.10
ENSMUST00000206124.2 |
Prss8
|
protease, serine 8 (prostasin) |
| chr7_-_25112256 | 7.32 |
ENSMUST00000200880.4
ENSMUST00000074040.4 |
Cxcl17
|
chemokine (C-X-C motif) ligand 17 |
| chr6_+_18170757 | 7.16 |
ENSMUST00000129452.8
|
Cftr
|
cystic fibrosis transmembrane conductance regulator |
| chr8_-_71219299 | 7.14 |
ENSMUST00000222087.4
|
Ifi30
|
interferon gamma inducible protein 30 |
| chr1_-_131204651 | 7.09 |
ENSMUST00000161764.8
|
Ikbke
|
inhibitor of kappaB kinase epsilon |
| chr9_+_92131797 | 7.07 |
ENSMUST00000093801.10
|
Plscr1
|
phospholipid scramblase 1 |
| chr8_+_22145796 | 6.93 |
ENSMUST00000079528.6
|
Defa17
|
defensin, alpha, 17 |
| chr4_-_58987094 | 6.79 |
ENSMUST00000030069.7
|
Ptgr1
|
prostaglandin reductase 1 |
| chrX_-_47551990 | 6.79 |
ENSMUST00000033429.9
ENSMUST00000140486.2 |
Elf4
|
E74-like factor 4 (ets domain transcription factor) |
| chr4_-_154384305 | 6.78 |
ENSMUST00000154895.2
|
Arhgef16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr1_+_171246593 | 6.77 |
ENSMUST00000171362.2
|
Tstd1
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1 |
| chr16_+_97157934 | 6.73 |
ENSMUST00000047275.8
|
Bace2
|
beta-site APP-cleaving enzyme 2 |
| chr8_+_105558204 | 6.71 |
ENSMUST00000059449.7
|
Ces2b
|
carboxyesterase 2B |
| chr5_-_145946408 | 6.70 |
ENSMUST00000138870.2
ENSMUST00000068317.13 |
Cyp3a25
|
cytochrome P450, family 3, subfamily a, polypeptide 25 |
| chr6_+_128639342 | 6.67 |
ENSMUST00000032518.7
ENSMUST00000204416.2 |
Clec2h
|
C-type lectin domain family 2, member h |
| chr7_+_43874752 | 6.67 |
ENSMUST00000075162.5
|
Klk1
|
kallikrein 1 |
| chr5_-_100710702 | 6.63 |
ENSMUST00000097437.9
|
Plac8
|
placenta-specific 8 |
| chr5_+_87148697 | 6.59 |
ENSMUST00000031186.9
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr6_+_138118565 | 6.53 |
ENSMUST00000118091.8
|
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr11_+_76792977 | 6.53 |
ENSMUST00000102495.8
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr1_+_88139678 | 6.42 |
ENSMUST00000073049.7
|
Ugt1a1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 |
| chr2_+_118603247 | 6.40 |
ENSMUST00000061360.4
ENSMUST00000130293.8 |
Phgr1
|
proline/histidine/glycine-rich 1 |
| chr19_+_39049442 | 6.32 |
ENSMUST00000087236.5
|
Cyp2c65
|
cytochrome P450, family 2, subfamily c, polypeptide 65 |
| chr17_+_48037758 | 6.21 |
ENSMUST00000024782.12
ENSMUST00000144955.2 |
Pgc
|
progastricsin (pepsinogen C) |
| chr16_-_56616186 | 6.20 |
ENSMUST00000023437.5
|
Adgrg7
|
adhesion G protein-coupled receptor G7 |
| chr8_+_21681630 | 6.19 |
ENSMUST00000098896.5
|
Defa31
|
defensin, alpha, 31 |
| chr11_+_69857722 | 6.17 |
ENSMUST00000151515.2
|
Cldn7
|
claudin 7 |
| chr7_+_99827886 | 6.16 |
ENSMUST00000207358.2
ENSMUST00000207995.2 ENSMUST00000049333.13 ENSMUST00000170954.10 ENSMUST00000179842.3 ENSMUST00000208260.2 |
Kcne3
|
potassium voltage-gated channel, Isk-related subfamily, gene 3 |
| chr2_-_65955338 | 6.14 |
ENSMUST00000028378.4
|
Galnt3
|
polypeptide N-acetylgalactosaminyltransferase 3 |
| chr6_-_87327885 | 6.10 |
ENSMUST00000032129.3
|
Gkn1
|
gastrokine 1 |
| chr9_+_45230370 | 5.97 |
ENSMUST00000034597.8
|
Tmprss13
|
transmembrane protease, serine 13 |
| chr2_+_151414524 | 5.95 |
ENSMUST00000028950.9
|
Sdcbp2
|
syndecan binding protein (syntenin) 2 |
| chr1_+_106788873 | 5.92 |
ENSMUST00000086701.13
ENSMUST00000188745.3 ENSMUST00000112730.8 |
Serpinb5
|
serine (or cysteine) peptidase inhibitor, clade B, member 5 |
| chr6_+_68247469 | 5.91 |
ENSMUST00000103321.3
|
Igkv1-110
|
immunoglobulin kappa variable 1-110 |
| chr5_+_35198853 | 5.87 |
ENSMUST00000030985.10
ENSMUST00000202573.2 |
Hgfac
|
hepatocyte growth factor activator |
| chr6_-_3487369 | 5.80 |
ENSMUST00000201607.4
|
Hepacam2
|
HEPACAM family member 2 |
| chr12_+_8062331 | 5.78 |
ENSMUST00000171239.2
|
Apob
|
apolipoprotein B |
| chr19_+_56276343 | 5.75 |
ENSMUST00000095948.11
|
Habp2
|
hyaluronic acid binding protein 2 |
| chr1_+_87983099 | 5.70 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr7_+_140415170 | 5.64 |
ENSMUST00000211372.2
ENSMUST00000026554.11 ENSMUST00000185612.3 |
Urah
|
urate (5-hydroxyiso-) hydrolase |
| chr1_-_155688551 | 5.62 |
ENSMUST00000194632.2
ENSMUST00000111764.8 |
Qsox1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr9_+_44953723 | 5.56 |
ENSMUST00000034600.5
|
Mpzl2
|
myelin protein zero-like 2 |
| chr15_+_10952418 | 5.50 |
ENSMUST00000022853.15
ENSMUST00000110523.2 |
C1qtnf3
|
C1q and tumor necrosis factor related protein 3 |
| chr4_+_152243324 | 5.48 |
ENSMUST00000030782.2
|
Hes2
|
hes family bHLH transcription factor 2 |
| chr4_-_63965161 | 5.47 |
ENSMUST00000107377.10
|
Tnc
|
tenascin C |
| chr13_-_100589357 | 5.44 |
ENSMUST00000222155.2
ENSMUST00000221727.2 |
Naip1
|
NLR family, apoptosis inhibitory protein 1 |
| chr2_+_5850053 | 5.42 |
ENSMUST00000127116.7
ENSMUST00000194933.2 |
Nudt5
|
nudix (nucleoside diphosphate linked moiety X)-type motif 5 |
| chr6_+_48881913 | 5.39 |
ENSMUST00000162948.7
ENSMUST00000167529.3 |
Aoc1
|
amine oxidase, copper-containing 1 |
| chr11_-_120971984 | 5.37 |
ENSMUST00000026162.12
|
Sectm1a
|
secreted and transmembrane 1A |
| chr17_-_37178079 | 5.33 |
ENSMUST00000025329.13
ENSMUST00000174195.8 |
Trim15
|
tripartite motif-containing 15 |
| chr4_-_49473904 | 5.32 |
ENSMUST00000135976.2
|
Acnat1
|
acyl-coenzyme A amino acid N-acyltransferase 1 |
| chr7_-_3680530 | 5.31 |
ENSMUST00000038743.15
|
Tmc4
|
transmembrane channel-like gene family 4 |
| chr1_+_131566223 | 5.29 |
ENSMUST00000112411.2
|
Ctse
|
cathepsin E |
| chr2_+_122607157 | 5.23 |
ENSMUST00000005953.11
|
Sqor
|
sulfide quinone oxidoreductase |
| chrX_-_111608339 | 5.22 |
ENSMUST00000039887.4
|
Pof1b
|
premature ovarian failure 1B |
| chr4_+_137180565 | 5.21 |
ENSMUST00000048893.4
|
1700013G24Rik
|
RIKEN cDNA 1700013G24 gene |
| chr19_+_39102342 | 5.14 |
ENSMUST00000087234.3
|
Cyp2c66
|
cytochrome P450, family 2, subfamily c, polypeptide 66 |
| chr11_-_78313043 | 5.14 |
ENSMUST00000001122.6
|
Slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr5_-_73789764 | 5.14 |
ENSMUST00000087177.4
|
Lrrc66
|
leucine rich repeat containing 66 |
| chr7_+_30399208 | 5.11 |
ENSMUST00000013227.8
|
2200002J24Rik
|
RIKEN cDNA 2200002J24 gene |
| chr3_-_92758591 | 5.05 |
ENSMUST00000054426.5
|
Lce1l
|
late cornified envelope 1L |
| chr13_-_42000958 | 5.05 |
ENSMUST00000072012.10
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr15_-_82223001 | 5.03 |
ENSMUST00000023088.8
|
Naga
|
N-acetyl galactosaminidase, alpha |
| chr14_-_63482668 | 5.03 |
ENSMUST00000118022.8
|
Gata4
|
GATA binding protein 4 |
| chr2_-_163592127 | 5.01 |
ENSMUST00000017841.4
|
Ada
|
adenosine deaminase |
| chr9_+_88209250 | 5.00 |
ENSMUST00000034992.8
|
Nt5e
|
5' nucleotidase, ecto |
| chr2_+_122607297 | 4.96 |
ENSMUST00000124460.2
ENSMUST00000147475.2 |
Sqor
|
sulfide quinone oxidoreductase |
| chr12_-_113223839 | 4.94 |
ENSMUST00000194738.6
ENSMUST00000178282.3 |
Igha
|
immunoglobulin heavy constant alpha |
| chr6_+_68233361 | 4.94 |
ENSMUST00000103320.3
|
Igkv14-111
|
immunoglobulin kappa variable 14-111 |
| chr15_+_10224052 | 4.93 |
ENSMUST00000128450.8
ENSMUST00000148257.8 ENSMUST00000128921.8 |
Prlr
|
prolactin receptor |
| chr9_-_22042930 | 4.92 |
ENSMUST00000213815.2
|
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr13_+_3887757 | 4.91 |
ENSMUST00000042219.6
|
Calm4
|
calmodulin 4 |
| chr19_-_34143437 | 4.89 |
ENSMUST00000025686.9
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr17_+_32877851 | 4.88 |
ENSMUST00000235086.2
|
Cyp4f40
|
cytochrome P450, family 4, subfamily f, polypeptide 40 |
| chr16_-_21980200 | 4.88 |
ENSMUST00000115379.2
|
Igf2bp2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr15_+_77361241 | 4.86 |
ENSMUST00000060551.9
|
Apol10a
|
apolipoprotein L 10A |
| chr19_-_40260060 | 4.85 |
ENSMUST00000068439.13
|
Pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr17_-_34694911 | 4.85 |
ENSMUST00000065841.5
|
Btnl4
|
butyrophilin-like 4 |
| chr14_-_63482720 | 4.78 |
ENSMUST00000067417.10
|
Gata4
|
GATA binding protein 4 |
| chr14_+_47605208 | 4.77 |
ENSMUST00000151405.9
|
Lgals3
|
lectin, galactose binding, soluble 3 |
| chr16_+_22737227 | 4.76 |
ENSMUST00000231880.2
|
Fetub
|
fetuin beta |
| chr16_-_18904240 | 4.75 |
ENSMUST00000103746.3
|
Iglv1
|
immunoglobulin lambda variable 1 |
| chr8_-_94063823 | 4.75 |
ENSMUST00000044602.8
|
Ces1g
|
carboxylesterase 1G |
| chr1_-_155688635 | 4.74 |
ENSMUST00000035325.15
|
Qsox1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr1_-_136158027 | 4.72 |
ENSMUST00000150163.8
ENSMUST00000144464.7 |
Inava
|
innate immunity activator |
| chr15_+_98468885 | 4.72 |
ENSMUST00000023728.8
|
Tex49
|
testis expressed 49 |
| chr11_+_96173475 | 4.71 |
ENSMUST00000168043.2
|
Hoxb8
|
homeobox B8 |
| chr10_+_79656823 | 4.70 |
ENSMUST00000169041.9
|
Misp
|
mitotic spindle positioning |
| chr6_-_68681962 | 4.69 |
ENSMUST00000103330.2
|
Igkv10-94
|
immunoglobulin kappa variable 10-94 |
| chr11_-_83421333 | 4.66 |
ENSMUST00000035938.3
|
Ccl5
|
chemokine (C-C motif) ligand 5 |
| chr7_-_18757461 | 4.65 |
ENSMUST00000036018.6
|
Foxa3
|
forkhead box A3 |
| chr4_+_118285275 | 4.64 |
ENSMUST00000006557.13
ENSMUST00000167636.8 ENSMUST00000102673.11 |
Elovl1
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 |
| chr11_-_75329726 | 4.59 |
ENSMUST00000108437.8
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr4_+_118384426 | 4.58 |
ENSMUST00000030261.6
|
2610528J11Rik
|
RIKEN cDNA 2610528J11 gene |
| chr7_-_140856642 | 4.50 |
ENSMUST00000080654.7
ENSMUST00000167263.9 |
Cdhr5
|
cadherin-related family member 5 |
| chr6_+_78357683 | 4.47 |
ENSMUST00000101272.3
|
Reg3a
|
regenerating islet-derived 3 alpha |
| chr11_+_94827050 | 4.47 |
ENSMUST00000001547.8
|
Col1a1
|
collagen, type I, alpha 1 |
| chr19_-_43879031 | 4.46 |
ENSMUST00000212048.2
|
Dnmbp
|
dynamin binding protein |
| chr2_+_27567213 | 4.43 |
ENSMUST00000077257.12
|
Rxra
|
retinoid X receptor alpha |
| chr16_+_22737128 | 4.42 |
ENSMUST00000170805.9
|
Fetub
|
fetuin beta |
| chr3_+_89136353 | 4.40 |
ENSMUST00000041142.4
|
Muc1
|
mucin 1, transmembrane |
| chr6_+_34575435 | 4.39 |
ENSMUST00000079391.10
ENSMUST00000142512.8 ENSMUST00000115027.8 ENSMUST00000115026.8 |
Cald1
|
caldesmon 1 |
| chr17_-_23964807 | 4.39 |
ENSMUST00000046525.10
|
Kremen2
|
kringle containing transmembrane protein 2 |
| chr17_+_23883974 | 4.37 |
ENSMUST00000233541.2
|
Bicdl2
|
BICD family like cargo adaptor 2 |
| chr10_+_79658392 | 4.36 |
ENSMUST00000219305.2
ENSMUST00000046833.5 ENSMUST00000218687.2 |
Misp
|
mitotic spindle positioning |
| chr13_-_41981812 | 4.34 |
ENSMUST00000223337.2
ENSMUST00000221691.2 |
Adtrp
|
androgen dependent TFPI regulating protein |
| chr17_-_12894716 | 4.33 |
ENSMUST00000024596.10
|
Slc22a1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr19_-_40260286 | 4.33 |
ENSMUST00000182432.2
|
Pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr6_-_131290790 | 4.32 |
ENSMUST00000049150.8
|
Styk1
|
serine/threonine/tyrosine kinase 1 |
| chr8_-_11528615 | 4.29 |
ENSMUST00000033900.7
|
Rab20
|
RAB20, member RAS oncogene family |
| chr6_+_41331039 | 4.29 |
ENSMUST00000072103.7
|
Try10
|
trypsin 10 |
| chr7_+_121464254 | 4.28 |
ENSMUST00000033161.7
|
Scnn1b
|
sodium channel, nonvoltage-gated 1 beta |
| chr5_-_66308421 | 4.28 |
ENSMUST00000200775.4
ENSMUST00000094756.11 |
Rbm47
|
RNA binding motif protein 47 |
| chr7_+_140414837 | 4.27 |
ENSMUST00000106050.8
|
Urah
|
urate (5-hydroxyiso-) hydrolase |
| chr7_+_28136861 | 4.27 |
ENSMUST00000108292.9
ENSMUST00000108289.8 |
Gmfg
|
glia maturation factor, gamma |
| chr11_+_95915366 | 4.24 |
ENSMUST00000103157.10
|
Gip
|
gastric inhibitory polypeptide |
| chr3_+_63202687 | 4.23 |
ENSMUST00000194836.6
ENSMUST00000191633.6 |
Mme
|
membrane metallo endopeptidase |
| chr16_-_38115172 | 4.21 |
ENSMUST00000023504.5
|
Nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr6_-_68713748 | 4.20 |
ENSMUST00000183936.2
ENSMUST00000196863.2 |
Igkv19-93
|
immunoglobulin kappa chain variable 19-93 |
| chr11_+_96173355 | 4.19 |
ENSMUST00000125410.2
|
Hoxb8
|
homeobox B8 |
| chr10_-_75673175 | 4.15 |
ENSMUST00000220440.2
|
Gstt2
|
glutathione S-transferase, theta 2 |
| chr10_-_22696025 | 4.15 |
ENSMUST00000218002.2
ENSMUST00000049930.9 |
Tcf21
|
transcription factor 21 |
| chr18_+_20691095 | 4.15 |
ENSMUST00000059787.15
ENSMUST00000120102.8 |
Dsg2
|
desmoglein 2 |
| chr12_-_114252202 | 4.12 |
ENSMUST00000195124.6
ENSMUST00000103481.3 |
Ighv3-6
|
immunoglobulin heavy variable 3-6 |
| chr12_-_40184174 | 4.09 |
ENSMUST00000078481.14
ENSMUST00000002640.6 |
Scin
|
scinderin |
| chr15_-_82223065 | 4.07 |
ENSMUST00000229733.2
ENSMUST00000229388.2 |
Naga
|
N-acetyl galactosaminidase, alpha |
| chr12_+_31440842 | 4.05 |
ENSMUST00000167432.8
|
Slc26a3
|
solute carrier family 26, member 3 |
| chr6_+_78402956 | 4.04 |
ENSMUST00000079926.6
|
Reg1
|
regenerating islet-derived 1 |
| chr7_+_140918793 | 4.04 |
ENSMUST00000026577.13
|
Eps8l2
|
EPS8-like 2 |
| chr12_-_114104740 | 4.03 |
ENSMUST00000103473.2
|
Ighv9-3
|
immunoglobulin heavy variable V9-3 |
| chr11_+_75546989 | 3.99 |
ENSMUST00000136935.2
|
Myo1c
|
myosin IC |
| chr4_-_3938352 | 3.96 |
ENSMUST00000003369.10
|
Plag1
|
pleiomorphic adenoma gene 1 |
| chr11_-_99519057 | 3.96 |
ENSMUST00000081007.7
|
Krtap4-1
|
keratin associated protein 4-1 |
| chr8_+_120163857 | 3.95 |
ENSMUST00000152420.8
ENSMUST00000212112.2 ENSMUST00000098365.4 |
Osgin1
|
oxidative stress induced growth inhibitor 1 |
| chr18_+_11790409 | 3.91 |
ENSMUST00000047322.8
|
Rbbp8
|
retinoblastoma binding protein 8, endonuclease |
| chr15_-_82222971 | 3.91 |
ENSMUST00000230380.2
|
Naga
|
N-acetyl galactosaminidase, alpha |
| chr13_+_74554509 | 3.90 |
ENSMUST00000222435.2
|
Ftl1-ps1
|
ferritin light polypeptide 1, pseudogene 1 |
| chr4_+_128887017 | 3.88 |
ENSMUST00000030583.13
ENSMUST00000102604.11 |
Ak2
|
adenylate kinase 2 |
| chr13_-_42001102 | 3.87 |
ENSMUST00000121404.8
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr14_-_63509131 | 3.84 |
ENSMUST00000132122.2
|
Gata4
|
GATA binding protein 4 |
| chr1_+_58841808 | 3.83 |
ENSMUST00000190213.2
|
Casp8
|
caspase 8 |
| chr13_-_41981893 | 3.83 |
ENSMUST00000137905.2
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr2_-_164198427 | 3.83 |
ENSMUST00000109367.10
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr7_+_43361930 | 3.82 |
ENSMUST00000066834.8
|
Klk13
|
kallikrein related-peptidase 13 |
| chr4_+_109272828 | 3.82 |
ENSMUST00000106618.8
|
Ttc39a
|
tetratricopeptide repeat domain 39A |
| chr17_+_35482063 | 3.81 |
ENSMUST00000172503.3
|
H2-D1
|
histocompatibility 2, D region locus 1 |
| chr17_+_48761916 | 3.80 |
ENSMUST00000074574.13
|
Unc5cl
|
unc-5 family C-terminal like |
| chr7_-_30672824 | 3.79 |
ENSMUST00000147431.2
ENSMUST00000098553.11 ENSMUST00000108116.10 |
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr18_+_20691278 | 3.78 |
ENSMUST00000121837.2
|
Dsg2
|
desmoglein 2 |
| chr13_-_42001075 | 3.78 |
ENSMUST00000179758.8
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr1_+_87983189 | 3.77 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr9_-_71499628 | 3.76 |
ENSMUST00000093823.8
|
Myzap
|
myocardial zonula adherens protein |
| chr10_-_95252712 | 3.76 |
ENSMUST00000020215.16
|
Socs2
|
suppressor of cytokine signaling 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.9 | 29.6 | GO:1904446 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) positive regulation of establishment of Sertoli cell barrier(GO:1904446) negative regulation of type B pancreatic cell development(GO:2000077) |
| 6.4 | 19.3 | GO:0051977 | lysophospholipid transport(GO:0051977) |
| 4.6 | 13.7 | GO:0060540 | lung lobe formation(GO:0060464) diaphragm morphogenesis(GO:0060540) |
| 3.9 | 47.1 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 3.8 | 11.5 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 3.3 | 9.9 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 3.2 | 9.7 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 3.0 | 9.1 | GO:0035874 | amiloride transport(GO:0015898) cellular response to copper ion starvation(GO:0035874) response to azide(GO:0097184) cellular response to azide(GO:0097185) |
| 2.6 | 7.9 | GO:0003165 | Purkinje myocyte development(GO:0003165) |
| 2.5 | 7.5 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 2.4 | 9.5 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 2.2 | 8.6 | GO:0045415 | negative regulation of interleukin-8 biosynthetic process(GO:0045415) |
| 2.2 | 6.5 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 2.1 | 27.3 | GO:1903207 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 2.1 | 6.2 | GO:0090320 | regulation of chylomicron remnant clearance(GO:0090320) |
| 2.1 | 4.1 | GO:0060435 | bronchiole development(GO:0060435) |
| 2.0 | 10.1 | GO:0044849 | estrous cycle(GO:0044849) |
| 2.0 | 7.9 | GO:1902896 | terminal web assembly(GO:1902896) |
| 1.9 | 5.8 | GO:0009073 | tyrosine biosynthetic process(GO:0006571) aromatic amino acid family biosynthetic process(GO:0009073) aromatic amino acid family biosynthetic process, prephenate pathway(GO:0009095) |
| 1.9 | 3.9 | GO:0046060 | dATP metabolic process(GO:0046060) |
| 1.8 | 5.5 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 1.8 | 7.3 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 1.8 | 7.1 | GO:2000373 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 1.7 | 7.0 | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 1.6 | 6.5 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 1.6 | 4.9 | GO:0042374 | menaquinone metabolic process(GO:0009233) phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 1.6 | 1.6 | GO:1990009 | retinal cell apoptotic process(GO:1990009) |
| 1.6 | 4.8 | GO:0043387 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) |
| 1.6 | 19.0 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 1.6 | 12.6 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 1.5 | 6.2 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 1.5 | 10.5 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 1.5 | 1.5 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 1.4 | 4.3 | GO:0071846 | actin filament debranching(GO:0071846) |
| 1.4 | 1.4 | GO:0002436 | immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) |
| 1.4 | 9.5 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 1.3 | 4.0 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 1.3 | 7.9 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 1.3 | 2.6 | GO:2001148 | dipeptide transmembrane transport(GO:0035442) regulation of oligopeptide transport(GO:0090088) regulation of dipeptide transport(GO:0090089) positive regulation of oligopeptide transport(GO:2000878) positive regulation of dipeptide transport(GO:2000880) regulation of dipeptide transmembrane transport(GO:2001148) positive regulation of dipeptide transmembrane transport(GO:2001150) |
| 1.3 | 7.6 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 1.2 | 5.0 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 1.2 | 6.2 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 1.2 | 3.7 | GO:0070194 | synaptonemal complex disassembly(GO:0070194) |
| 1.2 | 6.1 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 1.2 | 4.7 | GO:0035697 | CD8-positive, alpha-beta T cell extravasation(GO:0035697) |
| 1.2 | 3.5 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 1.2 | 4.6 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 1.2 | 6.9 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 1.1 | 3.4 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 1.1 | 10.2 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 1.1 | 13.5 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 1.1 | 4.5 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 1.1 | 4.4 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 1.1 | 13.0 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 1.1 | 6.5 | GO:0036257 | multivesicular body organization(GO:0036257) |
| 1.1 | 5.4 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 1.0 | 10.4 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 1.0 | 5.9 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 1.0 | 7.9 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 1.0 | 2.9 | GO:1901642 | purine nucleoside transmembrane transport(GO:0015860) nucleoside transmembrane transport(GO:1901642) |
| 1.0 | 20.3 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 1.0 | 6.8 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 1.0 | 12.6 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 1.0 | 4.8 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 1.0 | 2.9 | GO:0036275 | response to 5-fluorouracil(GO:0036275) |
| 0.9 | 3.8 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.9 | 3.6 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.9 | 4.5 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.9 | 6.3 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.9 | 3.6 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.9 | 7.2 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.9 | 2.7 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.9 | 4.4 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.9 | 0.9 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) ventricular cardiac muscle cell development(GO:0055015) |
| 0.9 | 18.2 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.8 | 0.8 | GO:0070340 | detection of bacterial lipopeptide(GO:0070340) |
| 0.8 | 3.3 | GO:0002445 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) positive regulation of type IIa hypersensitivity(GO:0001798) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.8 | 1.7 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.8 | 10.5 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.8 | 4.8 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.8 | 2.4 | GO:1903028 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.8 | 4.0 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.8 | 3.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.8 | 10.2 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) |
| 0.8 | 3.1 | GO:0071727 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.8 | 4.6 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.8 | 6.1 | GO:0070162 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.7 | 3.0 | GO:0021502 | neural fold elevation formation(GO:0021502) |
| 0.7 | 0.7 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.7 | 5.8 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.7 | 2.2 | GO:0090164 | asymmetric Golgi ribbon formation(GO:0090164) |
| 0.7 | 4.3 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.7 | 5.0 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.7 | 4.2 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.7 | 2.1 | GO:1901738 | regulation of vitamin A metabolic process(GO:1901738) |
| 0.7 | 2.8 | GO:0034760 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 0.7 | 4.2 | GO:1903436 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.7 | 8.3 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.7 | 6.1 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.7 | 3.4 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.7 | 4.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.7 | 2.0 | GO:1904456 | negative regulation of neuronal action potential(GO:1904456) |
| 0.7 | 3.3 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.7 | 2.0 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.7 | 5.3 | GO:0003072 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.7 | 1.3 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.7 | 2.6 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.6 | 2.6 | GO:0010979 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) bile acid signaling pathway(GO:0038183) |
| 0.6 | 3.8 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.6 | 2.5 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.6 | 1.9 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.6 | 4.3 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.6 | 3.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.6 | 1.9 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.6 | 1.9 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.6 | 4.9 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.6 | 1.8 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.6 | 7.3 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) |
| 0.6 | 3.6 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.6 | 3.0 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.6 | 5.4 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.6 | 1.8 | GO:1990168 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.6 | 2.4 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.6 | 6.0 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.6 | 1.8 | GO:0051714 | regulation of cytolysis in other organism(GO:0051710) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.6 | 3.0 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.6 | 4.1 | GO:1900141 | regulation of oligodendrocyte apoptotic process(GO:1900141) negative regulation of oligodendrocyte apoptotic process(GO:1900142) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.6 | 2.9 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.6 | 0.6 | GO:0072098 | intermediate mesoderm development(GO:0048389) pattern specification involved in mesonephros development(GO:0061227) anterior/posterior pattern specification involved in kidney development(GO:0072098) |
| 0.6 | 2.3 | GO:0010986 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.6 | 12.0 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.6 | 1.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.6 | 4.0 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.6 | 0.6 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.6 | 6.7 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.5 | 3.3 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.5 | 1.6 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.5 | 1.6 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.5 | 1.6 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.5 | 5.7 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.5 | 3.6 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.5 | 6.7 | GO:0038089 | positive regulation of cell migration by vascular endothelial growth factor signaling pathway(GO:0038089) |
| 0.5 | 3.1 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.5 | 1.5 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.5 | 1.5 | GO:0061723 | glycophagy(GO:0061723) |
| 0.5 | 2.5 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.5 | 2.0 | GO:0002353 | plasma kallikrein-kinin cascade(GO:0002353) |
| 0.5 | 6.3 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.5 | 7.3 | GO:0070633 | transepithelial transport(GO:0070633) |
| 0.5 | 5.8 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.5 | 2.9 | GO:0035547 | interferon-beta secretion(GO:0035546) regulation of interferon-beta secretion(GO:0035547) positive regulation of interferon-beta secretion(GO:0035549) |
| 0.5 | 0.9 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.5 | 0.5 | GO:0002254 | kinin cascade(GO:0002254) |
| 0.5 | 12.1 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.5 | 3.7 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.5 | 1.8 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.5 | 3.6 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.4 | 0.9 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.4 | 1.8 | GO:0043096 | purine nucleobase salvage(GO:0043096) |
| 0.4 | 3.1 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.4 | 1.8 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.4 | 2.7 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.4 | 1.8 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.4 | 2.6 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.4 | 6.6 | GO:0015747 | urate transport(GO:0015747) |
| 0.4 | 3.9 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.4 | 3.0 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.4 | 1.3 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.4 | 1.3 | GO:0046832 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.4 | 1.3 | GO:0032827 | negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.4 | 0.9 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) |
| 0.4 | 1.3 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.4 | 1.7 | GO:0002030 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) |
| 0.4 | 1.7 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.4 | 5.0 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.4 | 1.7 | GO:0090282 | positive regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0090282) |
| 0.4 | 1.2 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.4 | 30.4 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.4 | 2.1 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.4 | 1.2 | GO:0010710 | regulation of collagen catabolic process(GO:0010710) |
| 0.4 | 4.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.4 | 2.9 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.4 | 1.6 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.4 | 1.6 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.4 | 1.2 | GO:0031104 | dendrite regeneration(GO:0031104) |
| 0.4 | 3.2 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.4 | 0.8 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.4 | 9.9 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.4 | 0.8 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.4 | 0.8 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.4 | 5.5 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.4 | 1.2 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.4 | 2.0 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.4 | 11.7 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.4 | 12.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.4 | 1.5 | GO:0010958 | regulation of amino acid import(GO:0010958) |
| 0.4 | 3.1 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.4 | 6.5 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.4 | 1.9 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.4 | 2.6 | GO:0002727 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.4 | 2.9 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.4 | 2.6 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.4 | 10.6 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.4 | 2.9 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.4 | 1.4 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.4 | 10.4 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.4 | 1.8 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.4 | 6.7 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.4 | 13.8 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.4 | 1.1 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.3 | 1.4 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.3 | 1.4 | GO:0051329 | interphase(GO:0051325) mitotic interphase(GO:0051329) |
| 0.3 | 1.4 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.3 | 1.7 | GO:0031437 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.3 | 1.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.3 | 4.0 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.3 | 1.7 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.3 | 2.0 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.3 | 3.0 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.3 | 1.7 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.3 | 2.0 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.3 | 7.3 | GO:0044146 | negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.3 | 2.0 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.3 | 2.6 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.3 | 1.6 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.3 | 1.0 | GO:1904092 | regulation of autophagic cell death(GO:1904092) negative regulation of autophagic cell death(GO:1904093) |
| 0.3 | 2.3 | GO:1902947 | regulation of tau-protein kinase activity(GO:1902947) |
| 0.3 | 5.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.3 | 0.6 | GO:0071554 | cell wall macromolecule catabolic process(GO:0016998) cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.3 | 3.1 | GO:0060051 | negative regulation of protein glycosylation(GO:0060051) |
| 0.3 | 1.2 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.3 | 1.2 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.3 | 3.4 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.3 | 0.9 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.3 | 2.7 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.3 | 8.2 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.3 | 1.5 | GO:0042412 | L-cysteine catabolic process to taurine(GO:0019452) taurine biosynthetic process(GO:0042412) |
| 0.3 | 10.0 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.3 | 2.1 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.3 | 6.7 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.3 | 2.0 | GO:0072126 | positive regulation of glomerular mesangial cell proliferation(GO:0072126) |
| 0.3 | 2.0 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) |
| 0.3 | 4.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.3 | 2.0 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.3 | 3.7 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.3 | 1.1 | GO:0060376 | positive regulation of mast cell differentiation(GO:0060376) |
| 0.3 | 36.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.3 | 0.8 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.3 | 0.6 | GO:0032763 | regulation of mast cell cytokine production(GO:0032763) |
| 0.3 | 0.8 | GO:0009629 | response to gravity(GO:0009629) |
| 0.3 | 1.4 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.3 | 3.6 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.3 | 3.2 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.3 | 0.5 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.3 | 8.4 | GO:0070266 | necroptotic process(GO:0070266) |
| 0.3 | 1.6 | GO:2000317 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.3 | 2.1 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.3 | 3.9 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.3 | 1.8 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.3 | 1.6 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 0.3 | 0.8 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.3 | 0.5 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.3 | 1.0 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.3 | 8.8 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.3 | 14.0 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.3 | 2.3 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.3 | 4.1 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.3 | 2.3 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.3 | 0.5 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 0.3 | 2.3 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.3 | 0.8 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.3 | 0.3 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.3 | 6.3 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.2 | 1.2 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.2 | 2.2 | GO:0006559 | L-phenylalanine catabolic process(GO:0006559) tyrosine catabolic process(GO:0006572) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.2 | 0.7 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.2 | 0.7 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.2 | 12.1 | GO:0050891 | multicellular organismal water homeostasis(GO:0050891) |
| 0.2 | 2.4 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.2 | 2.9 | GO:0034143 | regulation of toll-like receptor 4 signaling pathway(GO:0034143) |
| 0.2 | 1.2 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.2 | 0.9 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.2 | 0.9 | GO:0010286 | heat acclimation(GO:0010286) |
| 0.2 | 2.8 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.2 | 0.7 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.2 | 2.1 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.2 | 2.6 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.2 | 0.2 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.2 | 12.9 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.2 | 1.6 | GO:0097503 | sialylation(GO:0097503) |
| 0.2 | 2.5 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.2 | 0.5 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.2 | 1.8 | GO:0048012 | hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.2 | 2.0 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.2 | 1.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.2 | 2.2 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.2 | 0.4 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.2 | 2.0 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 | 1.3 | GO:0072092 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.2 | 1.5 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.2 | 0.6 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.2 | 3.6 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.2 | 12.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.2 | 1.9 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.2 | 10.2 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.2 | 2.0 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.2 | 2.2 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.2 | 1.8 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.2 | 1.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.2 | 1.6 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.2 | 2.9 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.2 | 2.7 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.2 | 1.0 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.2 | 0.8 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.2 | 1.9 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.2 | 8.5 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.2 | 2.2 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.2 | 1.3 | GO:0060174 | pulmonary myocardium development(GO:0003350) limb bud formation(GO:0060174) |
| 0.2 | 1.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.2 | 2.6 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.2 | 1.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.2 | 0.2 | GO:0002246 | wound healing involved in inflammatory response(GO:0002246) inflammatory response to wounding(GO:0090594) |
| 0.2 | 2.5 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.2 | 2.0 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.2 | 4.6 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.2 | 3.0 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.2 | 1.4 | GO:1902172 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.2 | 1.2 | GO:0060022 | hard palate development(GO:0060022) |
| 0.2 | 0.3 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.2 | 3.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 0.2 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.2 | 1.7 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.2 | 2.2 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.2 | 1.5 | GO:0002067 | glandular epithelial cell differentiation(GO:0002067) |
| 0.2 | 1.6 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.2 | 1.1 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.2 | 1.8 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.2 | 2.2 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.2 | 0.5 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.2 | 1.0 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.2 | 2.6 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.2 | 1.3 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.2 | 2.5 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.2 | 4.0 | GO:0043616 | keratinocyte proliferation(GO:0043616) |
| 0.2 | 0.9 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.2 | 1.7 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.2 | 1.9 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 1.6 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.2 | 34.0 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.2 | 2.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 2.6 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.2 | 2.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 1.1 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.2 | 1.5 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.2 | 2.9 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.2 | 0.6 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.2 | 0.5 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.2 | 0.5 | GO:0046618 | drug export(GO:0046618) |
| 0.1 | 7.9 | GO:0045776 | negative regulation of blood pressure(GO:0045776) |
| 0.1 | 0.6 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 5.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 4.4 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.1 | 2.8 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 1.8 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 1.2 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 1.3 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.1 | 0.3 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.1 | 2.6 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.1 | 2.4 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 | 1.9 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 9.1 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 4.0 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 3.9 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 1.7 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.1 | 1.1 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.1 | 1.7 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 0.4 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 0.3 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.1 | 0.5 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.1 | 3.5 | GO:0072662 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 3.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 0.5 | GO:0002551 | mast cell chemotaxis(GO:0002551) |
| 0.1 | 2.7 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.1 | 0.5 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.4 | GO:0015838 | amino-acid betaine transport(GO:0015838) |
| 0.1 | 0.5 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 1.2 | GO:0035635 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.1 | 1.6 | GO:0071028 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.1 | 1.3 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 0.7 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.1 | 1.6 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 5.0 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.1 | 0.7 | GO:0035624 | regulation of vascular smooth muscle contraction(GO:0003056) receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.1 | 0.2 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 0.1 | 1.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.3 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 1.8 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 0.5 | GO:0072355 | histone H3-T3 phosphorylation(GO:0072355) |
| 0.1 | 4.4 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 1.6 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.1 | 7.1 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.1 | 0.8 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 1.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.4 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.1 | 1.7 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 2.1 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.1 | 0.3 | GO:0019427 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.1 | 2.4 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.1 | 0.8 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.5 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.1 | 0.7 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 0.8 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.2 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.1 | 0.5 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.8 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 1.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 2.5 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 1.5 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 0.7 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.1 | 0.5 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 | 0.4 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 1.2 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 4.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 0.7 | GO:0019359 | NADP biosynthetic process(GO:0006741) nicotinamide nucleotide biosynthetic process(GO:0019359) |
| 0.1 | 0.6 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 3.0 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.7 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 1.5 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) |
| 0.1 | 2.2 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 0.6 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.1 | 0.6 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 1.2 | GO:0052646 | alditol phosphate metabolic process(GO:0052646) |
| 0.1 | 1.7 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 0.3 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 1.3 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.1 | 0.7 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.1 | 0.4 | GO:0060931 | sinoatrial node cell differentiation(GO:0060921) sinoatrial node cell development(GO:0060931) |
| 0.1 | 1.7 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.1 | 0.8 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 2.0 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 | 0.2 | GO:1900756 | protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.1 | 0.9 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 2.6 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 2.2 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.1 | 1.4 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.1 | 0.5 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 0.6 | GO:0071435 | potassium ion export(GO:0071435) |
| 0.1 | 1.8 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.1 | 1.0 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 3.0 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.1 | 0.1 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.1 | 1.4 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.1 | 0.6 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 0.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.2 | GO:1900365 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 | 1.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 0.3 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 1.5 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.1 | 0.6 | GO:0002446 | neutrophil mediated immunity(GO:0002446) |
| 0.1 | 0.3 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 1.5 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 0.3 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.1 | 5.6 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 1.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.2 | GO:0035638 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.1 | 1.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 6.4 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.1 | 1.5 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.4 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 | 0.7 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.1 | 0.2 | GO:0035854 | regulation of primitive erythrocyte differentiation(GO:0010725) basophil differentiation(GO:0030221) eosinophil fate commitment(GO:0035854) |
| 0.1 | 1.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 0.7 | GO:0002363 | T-helper cell lineage commitment(GO:0002295) alpha-beta T cell lineage commitment(GO:0002363) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) T-helper 17 cell lineage commitment(GO:0072540) |
| 0.1 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 1.0 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.1 | 0.7 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.1 | 0.8 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 2.1 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 2.9 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 0.7 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.1 | 1.5 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.8 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.1 | 0.8 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.1 | 0.3 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.4 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 1.7 | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) |
| 0.1 | 0.3 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.1 | 0.6 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.1 | 0.4 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.1 | 0.3 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 1.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 | 0.3 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 0.2 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.1 | 0.2 | GO:0071348 | response to interleukin-11(GO:0071105) cellular response to interleukin-11(GO:0071348) |
| 0.1 | 0.9 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.1 | 0.7 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.3 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 1.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.5 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.2 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.9 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.3 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.5 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.0 | 0.5 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 1.3 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.3 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 1.5 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 | 2.0 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.4 | GO:0033216 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.2 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) histone H3-K36 methylation(GO:0010452) |
| 0.0 | 1.8 | GO:0003281 | ventricular septum development(GO:0003281) |
| 0.0 | 0.8 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.0 | 0.6 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 4.1 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.9 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.1 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.0 | 0.7 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.2 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.4 | GO:0010715 | regulation of extracellular matrix disassembly(GO:0010715) |
| 0.0 | 0.4 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.5 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.2 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) |
| 0.0 | 0.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.1 | GO:0022601 | menstrual cycle phase(GO:0022601) |
| 0.0 | 0.9 | GO:0042308 | negative regulation of protein import into nucleus(GO:0042308) negative regulation of protein import(GO:1904590) |
| 0.0 | 0.3 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 0.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 1.3 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 1.5 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.4 | GO:0021924 | cell proliferation in hindbrain(GO:0021534) cell proliferation in external granule layer(GO:0021924) cerebellar granule cell precursor proliferation(GO:0021930) |
| 0.0 | 0.2 | GO:0098779 | mitophagy in response to mitochondrial depolarization(GO:0098779) |
| 0.0 | 0.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.3 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 1.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.9 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.2 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 1.6 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.0 | 0.9 | GO:0042089 | regulation of cytokine biosynthetic process(GO:0042035) cytokine biosynthetic process(GO:0042089) |
| 0.0 | 1.2 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.6 | GO:0001825 | blastocyst formation(GO:0001825) |
| 0.0 | 0.4 | GO:0000578 | embryonic axis specification(GO:0000578) |
| 0.0 | 0.3 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 2.4 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.1 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.0 | 0.2 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 0.4 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.3 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.9 | GO:0048538 | thymus development(GO:0048538) |
| 0.0 | 1.7 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.4 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.6 | GO:1901655 | cellular response to ketone(GO:1901655) |
| 0.0 | 0.2 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.6 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 1.1 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0071354 | response to interleukin-6(GO:0070741) cellular response to interleukin-6(GO:0071354) |
| 0.0 | 0.1 | GO:0036296 | cellular response to increased oxygen levels(GO:0036295) response to increased oxygen levels(GO:0036296) |
| 0.0 | 0.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.0 | 0.4 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.1 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.3 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.0 | 0.2 | GO:0070228 | regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.0 | 0.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.3 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 29.6 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 2.0 | 6.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 1.9 | 5.8 | GO:0034359 | mature chylomicron(GO:0034359) |
| 1.9 | 9.6 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 1.6 | 6.3 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 1.5 | 4.5 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 1.5 | 7.4 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 1.4 | 4.3 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 1.4 | 8.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 1.3 | 5.4 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 1.3 | 6.3 | GO:0045160 | myosin I complex(GO:0045160) |
| 1.2 | 16.7 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 1.0 | 9.0 | GO:1990357 | terminal web(GO:1990357) |
| 1.0 | 2.9 | GO:0097132 | cyclin D2-CDK6 complex(GO:0097132) |
| 0.9 | 29.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.9 | 6.1 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.8 | 7.5 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.8 | 31.8 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.8 | 6.5 | GO:0042825 | TAP complex(GO:0042825) |
| 0.8 | 6.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.8 | 2.3 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.8 | 7.7 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.8 | 10.5 | GO:0043219 | lateral loop(GO:0043219) |
| 0.7 | 8.0 | GO:0045179 | apical cortex(GO:0045179) |
| 0.7 | 12.8 | GO:0090543 | Flemming body(GO:0090543) |
| 0.7 | 2.8 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.7 | 2.7 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.6 | 21.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.6 | 9.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.6 | 4.5 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.5 | 5.4 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.5 | 3.8 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.5 | 4.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.5 | 3.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.5 | 3.4 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.5 | 7.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.5 | 8.9 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.5 | 3.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.5 | 3.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.5 | 15.1 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.4 | 4.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.4 | 2.6 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.4 | 6.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.4 | 12.4 | GO:0044216 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.4 | 1.7 | GO:0008623 | CHRAC(GO:0008623) |
| 0.4 | 3.7 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.4 | 4.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.4 | 3.6 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.4 | 3.7 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.4 | 19.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.4 | 44.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.3 | 4.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.3 | 1.7 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.3 | 1.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.3 | 2.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.3 | 3.9 | GO:0005818 | aster(GO:0005818) |
| 0.3 | 3.9 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.3 | 7.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 19.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.3 | 2.6 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.3 | 2.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.3 | 39.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.3 | 8.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.3 | 1.5 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.3 | 3.0 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.3 | 3.0 | GO:0044815 | DNA packaging complex(GO:0044815) |
| 0.3 | 3.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 1.0 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.2 | 1.2 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.2 | 13.5 | GO:0031228 | intrinsic component of Golgi membrane(GO:0031228) |
| 0.2 | 0.9 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.2 | 3.8 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.2 | 1.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 3.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 0.4 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.2 | 1.5 | GO:0097413 | Lewy body(GO:0097413) |
| 0.2 | 3.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.2 | 0.7 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.2 | 0.5 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.2 | 1.3 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.2 | 3.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.2 | 0.5 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.2 | 0.2 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.2 | 0.4 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.2 | 4.0 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.2 | 1.4 | GO:0044299 | C-fiber(GO:0044299) |
| 0.2 | 1.7 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 0.7 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.2 | 1.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.2 | 2.9 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.2 | 1.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.2 | 1.5 | GO:0002177 | manchette(GO:0002177) |
| 0.2 | 2.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 2.5 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 7.6 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 2.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 14.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.0 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 2.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 0.4 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.1 | 0.7 | GO:0035841 | growing cell tip(GO:0035838) new growing cell tip(GO:0035841) |
| 0.1 | 10.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 5.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.9 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.8 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 1.3 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 11.8 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 0.5 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 9.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 0.4 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.1 | 10.1 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 10.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 2.0 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 2.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 1.4 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 4.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 2.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 1.1 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 1.0 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 1.9 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 1.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 8.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 3.4 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.1 | 14.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 0.6 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 182.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 1.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 2.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 1.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 1.9 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 2.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 6.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 0.4 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 0.9 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 1.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 0.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 1.7 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 3.0 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.6 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.1 | 2.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.5 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 3.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 12.7 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 9.4 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 6.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.4 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.7 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.7 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.4 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 3.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.8 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 1.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 6.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 2.3 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.1 | 1.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.4 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 1.0 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 2.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 3.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.1 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 17.9 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 8.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.3 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 8.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 6.5 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 4.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.6 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.5 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.7 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 6.2 | GO:0070161 | adherens junction(GO:0005912) anchoring junction(GO:0070161) |
| 0.0 | 5.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.6 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.5 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 1.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 2.4 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 45.1 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.3 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.9 | 29.6 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 4.3 | 13.0 | GO:0008456 | alpha-N-acetylgalactosaminidase activity(GO:0008456) |
| 3.6 | 18.2 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 3.3 | 9.9 | GO:0033971 | hydroxyisourate hydrolase activity(GO:0033971) |
| 3.0 | 9.1 | GO:0052599 | diamine oxidase activity(GO:0052597) histamine oxidase activity(GO:0052598) methylputrescine oxidase activity(GO:0052599) propane-1,3-diamine oxidase activity(GO:0052600) |
| 2.7 | 8.0 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 2.6 | 10.4 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 2.3 | 7.0 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 2.2 | 2.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 2.2 | 6.5 | GO:0046980 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 2.1 | 12.8 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 2.1 | 10.4 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 2.0 | 6.1 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 2.0 | 80.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 1.9 | 9.7 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 1.8 | 7.3 | GO:0042008 | interleukin-18 receptor activity(GO:0042008) |
| 1.8 | 9.0 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 1.7 | 6.8 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 1.7 | 6.8 | GO:0004058 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 1.6 | 9.8 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 1.6 | 6.5 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 1.6 | 4.8 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 1.6 | 4.7 | GO:0031726 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) CCR1 chemokine receptor binding(GO:0031726) |
| 1.4 | 4.3 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 1.4 | 8.6 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 1.2 | 10.8 | GO:0016672 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) |
| 1.2 | 17.4 | GO:0046977 | TAP binding(GO:0046977) |
| 1.1 | 3.4 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 1.1 | 4.4 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 1.1 | 6.5 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 1.0 | 13.6 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 1.0 | 3.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 1.0 | 7.2 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 1.0 | 4.9 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 1.0 | 4.8 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 1.0 | 2.9 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.9 | 2.8 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.9 | 5.6 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.9 | 4.6 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.9 | 2.7 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.9 | 3.6 | GO:0019862 | IgA binding(GO:0019862) |
| 0.9 | 2.7 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 0.9 | 7.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.9 | 2.6 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.8 | 1.7 | GO:0005350 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.8 | 10.6 | GO:0035473 | lipase binding(GO:0035473) |
| 0.8 | 14.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.8 | 11.9 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.8 | 7.9 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.8 | 15.6 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.8 | 2.3 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.8 | 3.0 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.7 | 5.9 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.7 | 3.7 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.7 | 1.4 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.7 | 2.2 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.7 | 2.9 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.7 | 8.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.7 | 2.0 | GO:0016034 | maleylacetoacetate isomerase activity(GO:0016034) |
| 0.7 | 2.0 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.7 | 5.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.7 | 14.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.7 | 2.0 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.7 | 5.3 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.7 | 7.9 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.6 | 1.9 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.6 | 2.6 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.6 | 21.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.6 | 6.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.6 | 1.9 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.6 | 3.7 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.6 | 4.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.6 | 1.8 | GO:0002055 | adenine binding(GO:0002055) adenine phosphoribosyltransferase activity(GO:0003999) |
| 0.6 | 22.5 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.6 | 3.5 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.6 | 7.0 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.6 | 1.7 | GO:0035730 | S-nitrosoglutathione binding(GO:0035730) dinitrosyl-iron complex binding(GO:0035731) |
| 0.6 | 2.2 | GO:0001512 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.6 | 3.9 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.6 | 3.9 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.5 | 5.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.5 | 5.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.5 | 3.8 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.5 | 4.8 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.5 | 3.1 | GO:0016019 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.5 | 1.5 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.5 | 6.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.5 | 7.0 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.5 | 8.9 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.5 | 3.0 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.5 | 8.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.5 | 2.4 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.5 | 5.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.5 | 3.8 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.5 | 1.4 | GO:0019002 | GMP binding(GO:0019002) |
| 0.5 | 2.3 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.5 | 2.3 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.5 | 2.3 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.5 | 1.8 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.4 | 4.0 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.4 | 2.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.4 | 5.0 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.4 | 0.8 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.4 | 5.3 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.4 | 1.6 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.4 | 1.6 | GO:0005534 | galactose binding(GO:0005534) |
| 0.4 | 8.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.4 | 1.2 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.4 | 1.6 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.4 | 1.2 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.4 | 3.6 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.4 | 8.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.4 | 1.2 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.4 | 5.5 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.4 | 1.2 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.4 | 51.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.4 | 1.5 | GO:0033797 | selenate reductase activity(GO:0033797) |
| 0.4 | 2.6 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.4 | 2.6 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.4 | 2.2 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.4 | 1.5 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.4 | 10.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.4 | 13.5 | GO:0043236 | laminin binding(GO:0043236) |
| 0.3 | 1.4 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.3 | 7.5 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.3 | 1.7 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.3 | 8.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.3 | 2.3 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.3 | 2.6 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.3 | 4.9 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.3 | 77.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.3 | 2.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.3 | 1.6 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.3 | 1.6 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.3 | 1.6 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.3 | 1.9 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.3 | 6.0 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.3 | 0.9 | GO:0004686 | elongation factor-2 kinase activity(GO:0004686) |
| 0.3 | 2.5 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.3 | 3.4 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.3 | 4.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.3 | 8.8 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.3 | 1.5 | GO:2001070 | starch binding(GO:2001070) |
| 0.3 | 1.8 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.3 | 1.8 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.3 | 7.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.3 | 6.7 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.3 | 1.7 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.3 | 1.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.3 | 7.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.3 | 3.9 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.3 | 0.3 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.3 | 1.1 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.3 | 1.6 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.3 | 1.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.3 | 2.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.3 | 1.8 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.3 | 1.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.3 | 0.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.3 | 4.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.3 | 4.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.3 | 0.8 | GO:0004823 | glutamine-tRNA ligase activity(GO:0004819) leucine-tRNA ligase activity(GO:0004823) |
| 0.2 | 1.5 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 1.5 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.2 | 1.2 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.2 | 3.6 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.2 | 1.0 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.2 | 1.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.2 | 14.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.2 | 0.9 | GO:0008147 | structural constituent of bone(GO:0008147) |
| 0.2 | 0.7 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.2 | 1.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 1.6 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 1.6 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.2 | 2.0 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.2 | 2.9 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.2 | 2.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.2 | 0.9 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.2 | 3.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 3.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 1.5 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.2 | 4.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 11.8 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.2 | 1.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.2 | 2.5 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 1.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 5.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 7.0 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.2 | 1.2 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.2 | 1.6 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.2 | 0.2 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.2 | 3.7 | GO:0016918 | retinal binding(GO:0016918) |
| 0.2 | 3.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.2 | 1.9 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 1.6 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.2 | 1.4 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.2 | 2.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 0.7 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.2 | 3.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.2 | 25.4 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.2 | 2.6 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 1.2 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.2 | 0.9 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 0.2 | 26.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 14.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.2 | 2.0 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.2 | 0.5 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.2 | 0.3 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.2 | 3.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 2.2 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.2 | 0.5 | GO:0051747 | cytosine C-5 DNA demethylase activity(GO:0051747) |
| 0.2 | 3.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 0.5 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.2 | 0.5 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 0.2 | 2.0 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.2 | 2.9 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.2 | 4.7 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.2 | 19.7 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.1 | 10.5 | GO:0008238 | exopeptidase activity(GO:0008238) |
| 0.1 | 4.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.6 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 2.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 1.8 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 1.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.4 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.1 | 1.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 16.1 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 0.7 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 1.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 2.6 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 23.6 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.1 | 1.3 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 3.7 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 3.1 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 1.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 0.9 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.1 | 0.2 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.1 | 3.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 4.7 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.1 | 0.3 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 1.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.8 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 0.3 | GO:0004420 | hydroxymethylglutaryl-CoA reductase (NADPH) activity(GO:0004420) hydroxymethylglutaryl-CoA reductase activity(GO:0042282) |
| 0.1 | 2.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 7.6 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 0.6 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 1.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.0 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 1.9 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.3 | GO:0004904 | interferon receptor activity(GO:0004904) type I interferon receptor activity(GO:0004905) type I interferon binding(GO:0019962) |
| 0.1 | 0.9 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.1 | 3.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.7 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.1 | 1.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 1.0 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.3 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.1 | 0.3 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.1 | 0.5 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.1 | 0.6 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.1 | 1.0 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 5.9 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 0.4 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 0.8 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 0.8 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.1 | 0.6 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 1.4 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 0.4 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 1.1 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 1.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.3 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.1 | 1.2 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 0.6 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 1.7 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 1.4 | GO:0035174 | AMP-activated protein kinase activity(GO:0004679) histone serine kinase activity(GO:0035174) |
| 0.1 | 1.4 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) hexose transmembrane transporter activity(GO:0015149) |
| 0.1 | 1.8 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 1.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 2.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.4 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.1 | 1.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 2.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 17.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 2.2 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.8 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 3.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.7 | GO:0005186 | pheromone activity(GO:0005186) |
| 0.1 | 1.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.8 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 0.3 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 2.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 1.5 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 1.6 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 1.0 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 0.9 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 14.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 0.7 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 5.1 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 0.4 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 27.8 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.1 | 1.4 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 3.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 1.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 6.0 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 2.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 1.4 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.6 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 1.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 2.5 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.9 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 1.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.3 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.2 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.6 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.5 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 1.3 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.2 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.2 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 1.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.1 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 1.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 2.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.0 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 11.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.5 | 32.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.4 | 6.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.4 | 20.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.4 | 22.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.3 | 15.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.3 | 2.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 0.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.3 | 8.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.3 | 2.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.3 | 15.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 18.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.3 | 11.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 2.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.2 | 17.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 54.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 6.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.2 | 3.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 35.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 3.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 2.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.2 | 5.0 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.2 | 11.7 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.2 | 1.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 7.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.2 | 11.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 2.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 0.7 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 3.0 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 6.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 3.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 5.5 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 2.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 4.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 2.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 1.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 2.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 1.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 4.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 5.0 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 1.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 0.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 10.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 3.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 0.3 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 2.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 1.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 1.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 5.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 5.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 0.9 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 0.8 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 1.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 3.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 2.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.8 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 2.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 4.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.9 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.4 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 21.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 1.3 | 22.9 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 1.0 | 22.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.9 | 20.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.9 | 14.7 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.7 | 8.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.7 | 30.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.7 | 9.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.6 | 8.5 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.5 | 28.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.5 | 26.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.5 | 7.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.5 | 35.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.5 | 10.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.5 | 14.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.5 | 9.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.4 | 18.7 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.4 | 20.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.4 | 5.7 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.4 | 7.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.4 | 6.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.4 | 7.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.4 | 3.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.3 | 2.7 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.3 | 3.0 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.3 | 6.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.3 | 5.0 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.3 | 9.1 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.3 | 4.7 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.3 | 0.8 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.3 | 4.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.3 | 8.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 8.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 5.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 2.8 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.2 | 3.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 10.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 1.9 | REACTOME S PHASE | Genes involved in S Phase |
| 0.2 | 2.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 10.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 1.8 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.2 | 1.4 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.2 | 2.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.2 | 3.7 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.2 | 2.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 1.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 4.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 0.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 3.8 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 3.1 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 9.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 2.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 7.6 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.2 | 1.6 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 12.6 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.2 | 3.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 12.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.2 | 1.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 2.3 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 2.0 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.1 | 1.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 4.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 0.9 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 8.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 4.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 2.0 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.1 | 15.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 2.2 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.1 | 1.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 4.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 4.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 2.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 4.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 2.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.8 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 5.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 0.9 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 1.6 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.1 | 8.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 3.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 4.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 5.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 1.8 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.1 | 2.2 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.1 | 1.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 4.4 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 1.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 12.2 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 0.3 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.1 | 0.7 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 0.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 5.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 4.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.0 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 5.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.1 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 0.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 2.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.8 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 3.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.8 | REACTOME ACTIVATED TLR4 SIGNALLING | Genes involved in Activated TLR4 signalling |
| 0.0 | 0.3 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.6 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 1.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.3 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.5 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.1 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.7 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |