Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
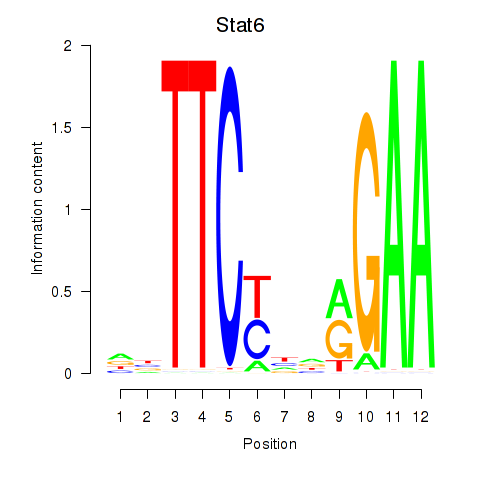
Results for Stat6
Z-value: 1.07
Transcription factors associated with Stat6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Stat6
|
ENSMUSG00000002147.19 | Stat6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Stat6 | mm39_v1_chr10_+_127478844_127478914 | 0.07 | 5.4e-01 | Click! |
Activity profile of Stat6 motif
Sorted Z-values of Stat6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Stat6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_8520006 | 7.04 |
ENSMUST00000162567.8
ENSMUST00000161217.8 |
Glcci1
|
glucocorticoid induced transcript 1 |
| chrX_-_60229164 | 6.96 |
ENSMUST00000166381.3
|
Cdr1
|
cerebellar degeneration related antigen 1 |
| chr10_+_126914755 | 5.43 |
ENSMUST00000039259.7
ENSMUST00000217941.2 |
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr6_-_78445846 | 4.79 |
ENSMUST00000032089.3
|
Reg3g
|
regenerating islet-derived 3 gamma |
| chr12_-_15866763 | 4.76 |
ENSMUST00000020922.8
ENSMUST00000221215.2 ENSMUST00000221518.2 |
Trib2
|
tribbles pseudokinase 2 |
| chr18_+_43897354 | 4.51 |
ENSMUST00000187157.7
ENSMUST00000043803.13 ENSMUST00000189750.2 |
Scgb3a2
|
secretoglobin, family 3A, member 2 |
| chr4_+_32982981 | 4.45 |
ENSMUST00000098190.10
ENSMUST00000029946.14 |
Rragd
|
Ras-related GTP binding D |
| chr11_+_97732108 | 4.36 |
ENSMUST00000155954.3
ENSMUST00000164364.2 ENSMUST00000170806.2 |
B230217C12Rik
|
RIKEN cDNA B230217C12 gene |
| chr11_+_75422516 | 4.24 |
ENSMUST00000149727.8
ENSMUST00000108433.8 ENSMUST00000042561.14 ENSMUST00000143035.8 |
Slc43a2
|
solute carrier family 43, member 2 |
| chr13_+_43938251 | 4.22 |
ENSMUST00000015540.4
|
Cd83
|
CD83 antigen |
| chr5_-_5315968 | 3.91 |
ENSMUST00000115451.8
ENSMUST00000115452.8 ENSMUST00000131392.8 |
Cdk14
|
cyclin-dependent kinase 14 |
| chr7_-_135130374 | 3.81 |
ENSMUST00000053716.8
|
Clrn3
|
clarin 3 |
| chr3_+_67799510 | 3.80 |
ENSMUST00000063263.5
ENSMUST00000182006.4 |
Iqcj
Iqschfp
|
IQ motif containing J Iqcj and Schip1 fusion protein |
| chr18_-_15536747 | 3.77 |
ENSMUST00000079081.8
|
Aqp4
|
aquaporin 4 |
| chr1_+_146373352 | 3.73 |
ENSMUST00000132847.8
ENSMUST00000166814.8 |
Brinp3
|
bone morphogenetic protein/retinoic acid inducible neural specific 3 |
| chr15_-_27788693 | 3.67 |
ENSMUST00000226287.2
|
Trio
|
triple functional domain (PTPRF interacting) |
| chr1_+_171594690 | 3.64 |
ENSMUST00000015460.5
|
Slamf1
|
signaling lymphocytic activation molecule family member 1 |
| chr11_+_75422925 | 3.58 |
ENSMUST00000169547.9
|
Slc43a2
|
solute carrier family 43, member 2 |
| chr3_-_87170903 | 3.53 |
ENSMUST00000090986.11
|
Fcrls
|
Fc receptor-like S, scavenger receptor |
| chr7_-_84021655 | 3.45 |
ENSMUST00000208392.2
ENSMUST00000208232.2 ENSMUST00000208863.2 |
Arnt2
|
aryl hydrocarbon receptor nuclear translocator 2 |
| chrX_+_162694397 | 3.42 |
ENSMUST00000140845.2
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr14_-_54648057 | 3.40 |
ENSMUST00000200545.2
ENSMUST00000227967.2 |
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr9_+_53757448 | 3.35 |
ENSMUST00000048485.7
|
Sln
|
sarcolipin |
| chr11_+_75422953 | 3.33 |
ENSMUST00000127226.3
|
Slc43a2
|
solute carrier family 43, member 2 |
| chr9_+_45029080 | 3.30 |
ENSMUST00000170998.9
ENSMUST00000093855.4 |
Scn2b
|
sodium channel, voltage-gated, type II, beta |
| chr6_+_71299758 | 3.26 |
ENSMUST00000065248.9
|
Cd8b1
|
CD8 antigen, beta chain 1 |
| chr5_-_71253107 | 3.11 |
ENSMUST00000197284.5
|
Gabra2
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 2 |
| chr5_+_8710059 | 2.93 |
ENSMUST00000047753.5
|
Abcb1a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1A |
| chr5_+_117501557 | 2.91 |
ENSMUST00000111959.2
|
Wsb2
|
WD repeat and SOCS box-containing 2 |
| chr1_+_134110142 | 2.89 |
ENSMUST00000082060.10
ENSMUST00000153856.8 ENSMUST00000133701.8 ENSMUST00000132873.8 |
Chil1
|
chitinase-like 1 |
| chr4_+_130202388 | 2.86 |
ENSMUST00000070532.8
|
Fabp3
|
fatty acid binding protein 3, muscle and heart |
| chr10_+_112107026 | 2.84 |
ENSMUST00000219301.2
ENSMUST00000092175.4 |
Kcnc2
|
potassium voltage gated channel, Shaw-related subfamily, member 2 |
| chr3_-_87171005 | 2.77 |
ENSMUST00000146512.2
|
Fcrls
|
Fc receptor-like S, scavenger receptor |
| chr5_+_57875309 | 2.76 |
ENSMUST00000191837.6
ENSMUST00000068110.10 |
Pcdh7
|
protocadherin 7 |
| chr1_-_46927230 | 2.71 |
ENSMUST00000185520.2
|
Slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr6_+_57679621 | 2.68 |
ENSMUST00000050077.15
|
Lancl2
|
LanC (bacterial lantibiotic synthetase component C)-like 2 |
| chr7_-_126399778 | 2.65 |
ENSMUST00000141355.4
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr14_-_56499690 | 2.65 |
ENSMUST00000015581.6
|
Gzmb
|
granzyme B |
| chr8_+_106331866 | 2.62 |
ENSMUST00000043531.10
|
Ripor1
|
RHO family interacting cell polarization regulator 1 |
| chr15_-_101778130 | 2.53 |
ENSMUST00000087996.7
|
Krt77
|
keratin 77 |
| chr6_+_48695535 | 2.49 |
ENSMUST00000127537.6
ENSMUST00000052503.8 |
Gm28053
Gimap7
|
predicted gene, 28053 GTPase, IMAP family member 7 |
| chr7_+_126690525 | 2.48 |
ENSMUST00000056288.7
ENSMUST00000206102.2 |
AI467606
|
expressed sequence AI467606 |
| chr6_-_87510200 | 2.41 |
ENSMUST00000113637.9
ENSMUST00000071024.7 |
Arhgap25
|
Rho GTPase activating protein 25 |
| chr7_+_87233554 | 2.39 |
ENSMUST00000125009.9
|
Grm5
|
glutamate receptor, metabotropic 5 |
| chr5_-_148329615 | 2.36 |
ENSMUST00000138257.8
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr1_+_134109888 | 2.25 |
ENSMUST00000156873.8
|
Chil1
|
chitinase-like 1 |
| chr6_+_38358404 | 2.25 |
ENSMUST00000162554.8
ENSMUST00000161751.8 |
Ttc26
|
tetratricopeptide repeat domain 26 |
| chr1_-_83016152 | 2.24 |
ENSMUST00000164473.2
ENSMUST00000045560.15 |
Slc19a3
|
solute carrier family 19, member 3 |
| chr7_-_126399208 | 2.24 |
ENSMUST00000133514.8
ENSMUST00000151137.8 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr7_-_57159743 | 2.17 |
ENSMUST00000068456.8
ENSMUST00000206734.2 |
Gabra5
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 5 |
| chr7_-_126399574 | 2.15 |
ENSMUST00000106348.8
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr1_-_46893206 | 2.13 |
ENSMUST00000027131.6
|
Slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr6_+_57679455 | 2.13 |
ENSMUST00000072954.8
|
Lancl2
|
LanC (bacterial lantibiotic synthetase component C)-like 2 |
| chr4_-_116982804 | 2.11 |
ENSMUST00000183310.2
|
Btbd19
|
BTB (POZ) domain containing 19 |
| chr13_-_41513215 | 2.08 |
ENSMUST00000224803.2
|
Nedd9
|
neural precursor cell expressed, developmentally down-regulated gene 9 |
| chr15_-_93417380 | 2.07 |
ENSMUST00000109255.3
|
Prickle1
|
prickle planar cell polarity protein 1 |
| chr2_+_163067283 | 2.04 |
ENSMUST00000099110.10
ENSMUST00000165937.8 |
Tox2
|
TOX high mobility group box family member 2 |
| chr2_+_70393782 | 2.01 |
ENSMUST00000123330.3
|
Gad1
|
glutamate decarboxylase 1 |
| chr14_+_61844899 | 2.01 |
ENSMUST00000225582.2
ENSMUST00000051184.10 |
Kcnrg
|
potassium channel regulator |
| chr6_+_41139948 | 1.98 |
ENSMUST00000103275.4
|
Trbv17
|
T cell receptor beta, variable 17 |
| chr19_-_7272758 | 1.98 |
ENSMUST00000025921.15
|
Mark2
|
MAP/microtubule affinity regulating kinase 2 |
| chr2_+_120331693 | 1.96 |
ENSMUST00000141181.9
|
Capn3
|
calpain 3 |
| chr13_-_77283534 | 1.92 |
ENSMUST00000159462.3
ENSMUST00000151524.9 |
Slf1
|
SMC5-SMC6 complex localization factor 1 |
| chr18_+_50186349 | 1.90 |
ENSMUST00000148159.3
|
Tnfaip8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr5_-_31265562 | 1.88 |
ENSMUST00000201396.2
ENSMUST00000202740.4 |
Slc30a3
|
solute carrier family 30 (zinc transporter), member 3 |
| chr5_+_114924106 | 1.87 |
ENSMUST00000137519.2
|
Ankrd13a
|
ankyrin repeat domain 13a |
| chr14_-_36641270 | 1.84 |
ENSMUST00000182797.8
|
Ccser2
|
coiled-coil serine rich 2 |
| chr1_+_160970276 | 1.83 |
ENSMUST00000111608.8
ENSMUST00000052245.9 |
Ankrd45
|
ankyrin repeat domain 45 |
| chr4_+_43406435 | 1.77 |
ENSMUST00000098106.9
ENSMUST00000139198.2 |
Rusc2
|
RUN and SH3 domain containing 2 |
| chr1_-_125840838 | 1.77 |
ENSMUST00000161361.3
|
Lypd1
|
Ly6/Plaur domain containing 1 |
| chr5_+_72360631 | 1.76 |
ENSMUST00000169617.3
|
Atp10d
|
ATPase, class V, type 10D |
| chr2_-_73284262 | 1.70 |
ENSMUST00000102679.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr1_+_179788675 | 1.69 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr5_+_14075281 | 1.67 |
ENSMUST00000073957.8
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr17_+_48606948 | 1.67 |
ENSMUST00000233092.2
|
Treml2
|
triggering receptor expressed on myeloid cells-like 2 |
| chr18_+_37827413 | 1.60 |
ENSMUST00000193414.2
|
Pcdhga5
|
protocadherin gamma subfamily A, 5 |
| chr19_+_3818112 | 1.58 |
ENSMUST00000005518.16
ENSMUST00000237440.2 ENSMUST00000152935.8 ENSMUST00000176262.8 ENSMUST00000176407.8 ENSMUST00000176926.8 ENSMUST00000176512.8 |
Kmt5b
|
lysine methyltransferase 5B |
| chr19_+_12647803 | 1.57 |
ENSMUST00000207341.3
ENSMUST00000208494.3 ENSMUST00000208657.3 |
Olfr1442
|
olfactory receptor 1442 |
| chr10_-_100425067 | 1.55 |
ENSMUST00000218821.2
ENSMUST00000054471.10 |
4930430F08Rik
|
RIKEN cDNA 4930430F08 gene |
| chr11_+_101136821 | 1.55 |
ENSMUST00000129680.8
|
Ramp2
|
receptor (calcitonin) activity modifying protein 2 |
| chr1_+_131936022 | 1.55 |
ENSMUST00000146432.2
|
Elk4
|
ELK4, member of ETS oncogene family |
| chr2_+_120331784 | 1.54 |
ENSMUST00000151342.3
|
Capn3
|
calpain 3 |
| chrX_+_149981074 | 1.52 |
ENSMUST00000184730.8
ENSMUST00000184392.8 ENSMUST00000096285.5 |
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chr6_+_48695571 | 1.52 |
ENSMUST00000204785.2
|
Gimap7
|
GTPase, IMAP family member 7 |
| chr7_+_80764547 | 1.51 |
ENSMUST00000026820.11
|
Slc28a1
|
solute carrier family 28 (sodium-coupled nucleoside transporter), member 1 |
| chr11_-_3143387 | 1.48 |
ENSMUST00000081318.13
ENSMUST00000142315.8 ENSMUST00000118627.2 ENSMUST00000066391.14 |
Sfi1
|
Sfi1 homolog, spindle assembly associated (yeast) |
| chr2_+_29236815 | 1.47 |
ENSMUST00000028139.11
ENSMUST00000113830.11 |
Med27
|
mediator complex subunit 27 |
| chrX_-_104919201 | 1.47 |
ENSMUST00000198209.2
|
Atrx
|
ATRX, chromatin remodeler |
| chr7_+_100435548 | 1.46 |
ENSMUST00000216021.2
|
Fam168a
|
family with sequence similarity 168, member A |
| chr6_+_149226891 | 1.46 |
ENSMUST00000189837.2
|
Resf1
|
retroelement silencing factor 1 |
| chr2_+_70393195 | 1.45 |
ENSMUST00000130998.8
|
Gad1
|
glutamate decarboxylase 1 |
| chr17_+_29309942 | 1.45 |
ENSMUST00000119901.9
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chr4_-_149747644 | 1.43 |
ENSMUST00000105689.8
|
Pik3cd
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta |
| chr7_+_80764564 | 1.41 |
ENSMUST00000119083.2
|
Slc28a1
|
solute carrier family 28 (sodium-coupled nucleoside transporter), member 1 |
| chr6_+_127430668 | 1.41 |
ENSMUST00000039680.7
|
Parp11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr11_-_3864664 | 1.40 |
ENSMUST00000109995.2
ENSMUST00000051207.2 |
Slc35e4
|
solute carrier family 35, member E4 |
| chr1_-_175520185 | 1.40 |
ENSMUST00000104984.4
ENSMUST00000209720.2 ENSMUST00000211489.2 ENSMUST00000210367.2 ENSMUST00000027809.8 |
Chml
Opn3
|
choroideremia-like opsin 3 |
| chr18_+_42669322 | 1.36 |
ENSMUST00000236418.2
|
Tcerg1
|
transcription elongation regulator 1 (CA150) |
| chr11_-_83421333 | 1.35 |
ENSMUST00000035938.3
|
Ccl5
|
chemokine (C-C motif) ligand 5 |
| chr6_+_127430630 | 1.35 |
ENSMUST00000112193.8
|
Parp11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr5_+_122239007 | 1.31 |
ENSMUST00000014080.13
ENSMUST00000111750.8 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr18_+_69654900 | 1.31 |
ENSMUST00000202057.4
|
Tcf4
|
transcription factor 4 |
| chr4_-_119349760 | 1.31 |
ENSMUST00000049994.8
|
Rimkla
|
ribosomal modification protein rimK-like family member A |
| chr3_+_90383425 | 1.29 |
ENSMUST00000001042.10
|
Ilf2
|
interleukin enhancer binding factor 2 |
| chr18_+_37488174 | 1.28 |
ENSMUST00000192867.2
ENSMUST00000051163.3 |
Pcdhb8
|
protocadherin beta 8 |
| chr3_-_33039225 | 1.27 |
ENSMUST00000108221.2
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr7_-_103666510 | 1.25 |
ENSMUST00000215653.2
|
Olfr639
|
olfactory receptor 639 |
| chr1_-_160134873 | 1.22 |
ENSMUST00000193185.6
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr9_-_49710190 | 1.21 |
ENSMUST00000114476.8
ENSMUST00000193547.6 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr5_-_125201872 | 1.21 |
ENSMUST00000055256.14
|
Ncor2
|
nuclear receptor co-repressor 2 |
| chr10_-_79940168 | 1.19 |
ENSMUST00000219260.2
|
Sbno2
|
strawberry notch 2 |
| chr6_+_34840057 | 1.17 |
ENSMUST00000074949.4
|
Tmem140
|
transmembrane protein 140 |
| chr7_-_28824440 | 1.17 |
ENSMUST00000214374.2
ENSMUST00000179893.9 ENSMUST00000032813.10 |
Ryr1
|
ryanodine receptor 1, skeletal muscle |
| chr7_+_97229065 | 1.15 |
ENSMUST00000107153.3
|
Rsf1
|
remodeling and spacing factor 1 |
| chr9_+_74959259 | 1.13 |
ENSMUST00000170310.2
ENSMUST00000166549.2 |
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr19_-_8690330 | 1.13 |
ENSMUST00000206598.2
|
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr1_-_23948764 | 1.12 |
ENSMUST00000129254.8
|
Smap1
|
small ArfGAP 1 |
| chr6_+_34840151 | 1.07 |
ENSMUST00000202010.2
|
Tmem140
|
transmembrane protein 140 |
| chr3_+_103739877 | 1.07 |
ENSMUST00000062945.12
|
Bcl2l15
|
BCLl2-like 15 |
| chr9_-_49710058 | 1.06 |
ENSMUST00000192584.2
ENSMUST00000166811.9 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr10_-_18891095 | 1.05 |
ENSMUST00000019997.11
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr3_-_144738526 | 1.04 |
ENSMUST00000029919.7
|
Clca1
|
chloride channel accessory 1 |
| chr17_+_48607405 | 1.04 |
ENSMUST00000170941.3
|
Treml2
|
triggering receptor expressed on myeloid cells-like 2 |
| chr9_+_74944994 | 1.02 |
ENSMUST00000007800.8
|
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr14_+_53913598 | 1.02 |
ENSMUST00000103662.6
|
Trav9-4
|
T cell receptor alpha variable 9-4 |
| chr5_+_122239030 | 1.01 |
ENSMUST00000139213.8
ENSMUST00000111751.8 ENSMUST00000155612.8 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr8_-_106427696 | 1.00 |
ENSMUST00000042608.8
|
Acd
|
adrenocortical dysplasia |
| chr2_+_69869459 | 0.99 |
ENSMUST00000060208.11
|
Myo3b
|
myosin IIIB |
| chr19_+_29229147 | 0.98 |
ENSMUST00000025705.7
ENSMUST00000065796.10 ENSMUST00000236990.2 |
Jak2
|
Janus kinase 2 |
| chr3_+_96484294 | 0.97 |
ENSMUST00000148290.2
|
Gm16253
|
predicted gene 16253 |
| chrX_+_133657312 | 0.97 |
ENSMUST00000081834.10
ENSMUST00000086880.11 ENSMUST00000086884.5 |
Armcx3
|
armadillo repeat containing, X-linked 3 |
| chr15_-_9529898 | 0.97 |
ENSMUST00000228782.2
ENSMUST00000003981.6 |
Il7r
|
interleukin 7 receptor |
| chr18_+_66591604 | 0.96 |
ENSMUST00000025399.9
ENSMUST00000237161.2 ENSMUST00000236933.2 |
Pmaip1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr18_-_36859732 | 0.96 |
ENSMUST00000061829.8
|
Cd14
|
CD14 antigen |
| chr7_-_97228589 | 0.96 |
ENSMUST00000151840.2
ENSMUST00000135998.8 ENSMUST00000144858.8 ENSMUST00000146605.8 ENSMUST00000072725.12 ENSMUST00000138060.3 ENSMUST00000154853.8 ENSMUST00000136757.8 ENSMUST00000124552.3 |
Aamdc
|
adipogenesis associated Mth938 domain containing |
| chr14_-_104760051 | 0.94 |
ENSMUST00000022716.4
ENSMUST00000228448.2 ENSMUST00000227640.2 |
Obi1
|
ORC ubiquitin ligase 1 |
| chr14_+_79825131 | 0.94 |
ENSMUST00000054908.10
|
Sugt1
|
SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) |
| chr1_+_173501215 | 0.94 |
ENSMUST00000085876.12
|
Ifi208
|
interferon activated gene 208 |
| chr7_+_6346723 | 0.93 |
ENSMUST00000207173.3
|
Gm3854
|
predicted gene 3854 |
| chr8_-_111808674 | 0.92 |
ENSMUST00000039597.14
|
Clec18a
|
C-type lectin domain family 18, member A |
| chr19_-_11852453 | 0.91 |
ENSMUST00000213954.2
ENSMUST00000217617.2 |
Olfr1419
|
olfactory receptor 1419 |
| chr1_+_160970233 | 0.91 |
ENSMUST00000125018.8
|
Ankrd45
|
ankyrin repeat domain 45 |
| chr7_+_107166925 | 0.90 |
ENSMUST00000239087.2
|
Olfml1
|
olfactomedin-like 1 |
| chr11_+_108477903 | 0.90 |
ENSMUST00000146912.9
|
Cep112
|
centrosomal protein 112 |
| chr10_-_78187524 | 0.89 |
ENSMUST00000166360.8
|
Agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr18_+_32970278 | 0.89 |
ENSMUST00000053663.11
|
Wdr36
|
WD repeat domain 36 |
| chr1_-_181670599 | 0.89 |
ENSMUST00000193030.6
|
Lbr
|
lamin B receptor |
| chr3_-_141637245 | 0.88 |
ENSMUST00000106232.8
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B |
| chr3_+_96939732 | 0.87 |
ENSMUST00000132256.8
ENSMUST00000072600.7 |
Gja5
|
gap junction protein, alpha 5 |
| chr2_-_148285450 | 0.87 |
ENSMUST00000099269.4
|
Cd93
|
CD93 antigen |
| chr11_-_55310724 | 0.86 |
ENSMUST00000108858.8
ENSMUST00000141530.2 |
Sparc
|
secreted acidic cysteine rich glycoprotein |
| chr7_-_3619132 | 0.81 |
ENSMUST00000039507.15
ENSMUST00000108645.8 ENSMUST00000148012.2 |
Oscar
|
osteoclast associated receptor |
| chr1_+_171668173 | 0.81 |
ENSMUST00000136479.8
|
Cd84
|
CD84 antigen |
| chr5_+_31212165 | 0.81 |
ENSMUST00000202795.4
ENSMUST00000201182.4 ENSMUST00000200953.4 |
Cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr11_-_61821023 | 0.79 |
ENSMUST00000108710.2
|
Akap10
|
A kinase (PRKA) anchor protein 10 |
| chr10_-_44334683 | 0.79 |
ENSMUST00000105490.3
|
Prdm1
|
PR domain containing 1, with ZNF domain |
| chr9_+_40092216 | 0.79 |
ENSMUST00000218134.2
ENSMUST00000216720.2 ENSMUST00000214763.2 |
Olfr986
|
olfactory receptor 986 |
| chr11_+_68393845 | 0.77 |
ENSMUST00000102613.8
ENSMUST00000060441.7 |
Pik3r6
|
phosphoinositide-3-kinase regulatory subunit 5 |
| chr14_+_53220658 | 0.73 |
ENSMUST00000200548.2
|
Trav9d-4
|
T cell receptor alpha variable 9D-4 |
| chr15_-_81810349 | 0.71 |
ENSMUST00000023113.7
|
Polr3h
|
polymerase (RNA) III (DNA directed) polypeptide H |
| chr5_-_39034039 | 0.70 |
ENSMUST00000169819.5
ENSMUST00000171633.5 |
Clnk
|
cytokine-dependent hematopoietic cell linker |
| chr11_-_61821072 | 0.69 |
ENSMUST00000102650.10
|
Akap10
|
A kinase (PRKA) anchor protein 10 |
| chr5_-_72893941 | 0.68 |
ENSMUST00000169534.6
|
Txk
|
TXK tyrosine kinase |
| chr2_+_118730823 | 0.67 |
ENSMUST00000151162.2
|
Bahd1
|
bromo adjacent homology domain containing 1 |
| chr1_-_157072060 | 0.67 |
ENSMUST00000129880.8
|
Rasal2
|
RAS protein activator like 2 |
| chr18_+_32970363 | 0.66 |
ENSMUST00000166214.9
|
Wdr36
|
WD repeat domain 36 |
| chr8_-_3976808 | 0.66 |
ENSMUST00000188386.2
ENSMUST00000084086.9 ENSMUST00000171635.8 |
Cd209b
|
CD209b antigen |
| chr8_+_106427774 | 0.65 |
ENSMUST00000098444.9
ENSMUST00000212352.2 |
Pard6a
|
par-6 family cell polarity regulator alpha |
| chr5_-_6926523 | 0.65 |
ENSMUST00000164784.2
|
Zfp804b
|
zinc finger protein 804B |
| chr9_-_96601574 | 0.64 |
ENSMUST00000128269.8
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chr2_+_31578537 | 0.64 |
ENSMUST00000075759.13
|
Abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr9_+_86625694 | 0.63 |
ENSMUST00000179574.2
ENSMUST00000036426.13 |
Prss35
|
protease, serine 35 |
| chr16_-_30152708 | 0.62 |
ENSMUST00000229503.2
|
Atp13a3
|
ATPase type 13A3 |
| chr4_-_41695442 | 0.62 |
ENSMUST00000102961.10
|
Cntfr
|
ciliary neurotrophic factor receptor |
| chr19_-_5974825 | 0.62 |
ENSMUST00000055458.6
|
Cdc42ep2
|
CDC42 effector protein (Rho GTPase binding) 2 |
| chr14_-_52277310 | 0.61 |
ENSMUST00000216907.2
ENSMUST00000214071.2 ENSMUST00000216188.2 |
Olfr221
|
olfactory receptor 221 |
| chr5_-_24650164 | 0.60 |
ENSMUST00000115043.8
ENSMUST00000115041.2 ENSMUST00000030800.13 |
Fastk
|
Fas-activated serine/threonine kinase |
| chrX_-_104918911 | 0.60 |
ENSMUST00000200471.2
|
Atrx
|
ATRX, chromatin remodeler |
| chr2_-_144392696 | 0.59 |
ENSMUST00000028915.6
|
Rbbp9
|
retinoblastoma binding protein 9, serine hydrolase |
| chr10_-_51507527 | 0.58 |
ENSMUST00000219286.2
ENSMUST00000020062.4 ENSMUST00000218684.2 |
Gprc6a
|
G protein-coupled receptor, family C, group 6, member A |
| chr5_-_103777145 | 0.57 |
ENSMUST00000031263.2
|
Slc10a6
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 6 |
| chr5_+_31212110 | 0.54 |
ENSMUST00000013773.12
ENSMUST00000201838.4 |
Cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr7_+_107166653 | 0.52 |
ENSMUST00000120990.2
|
Olfml1
|
olfactomedin-like 1 |
| chr7_+_127160751 | 0.52 |
ENSMUST00000190278.3
|
Tmem265
|
transmembrane protein 265 |
| chr5_+_102992873 | 0.51 |
ENSMUST00000070000.6
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr2_+_111329683 | 0.51 |
ENSMUST00000219064.3
|
Olfr1291-ps1
|
olfactory receptor 1291, pseudogene 1 |
| chr2_+_111136546 | 0.50 |
ENSMUST00000090329.2
|
Olfr1279
|
olfactory receptor 1279 |
| chr1_+_171668122 | 0.49 |
ENSMUST00000135386.2
|
Cd84
|
CD84 antigen |
| chr10_-_78187545 | 0.49 |
ENSMUST00000105390.8
|
Agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr15_+_51741138 | 0.48 |
ENSMUST00000136129.2
|
Utp23
|
UTP23 small subunit processome component |
| chr1_-_125839897 | 0.45 |
ENSMUST00000159417.2
|
Lypd1
|
Ly6/Plaur domain containing 1 |
| chr1_+_120048890 | 0.45 |
ENSMUST00000027637.13
ENSMUST00000112644.9 ENSMUST00000056038.15 |
3110009E18Rik
|
RIKEN cDNA 3110009E18 gene |
| chr7_-_104646109 | 0.42 |
ENSMUST00000050599.5
|
Olfr672
|
olfactory receptor 672 |
| chr11_+_78079243 | 0.40 |
ENSMUST00000002128.14
ENSMUST00000150941.8 |
Rab34
|
RAB34, member RAS oncogene family |
| chr17_-_36290129 | 0.38 |
ENSMUST00000165613.9
ENSMUST00000173872.8 |
Prr3
|
proline-rich polypeptide 3 |
| chr11_+_84771133 | 0.38 |
ENSMUST00000020837.7
|
Myo19
|
myosin XIX |
| chr15_+_6451721 | 0.38 |
ENSMUST00000163082.2
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr12_+_37158532 | 0.38 |
ENSMUST00000041183.7
|
Meox2
|
mesenchyme homeobox 2 |
| chrX_-_47551990 | 0.38 |
ENSMUST00000033429.9
ENSMUST00000140486.2 |
Elf4
|
E74-like factor 4 (ets domain transcription factor) |
| chr16_+_48877762 | 0.36 |
ENSMUST00000168680.2
|
Myh15
|
myosin, heavy chain 15 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 1.2 | 3.6 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 1.2 | 4.8 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.9 | 2.8 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.9 | 3.5 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.8 | 2.4 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.8 | 2.3 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.8 | 3.8 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.7 | 2.9 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.7 | 2.9 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.7 | 2.9 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.7 | 2.1 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.7 | 3.3 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.7 | 2.6 | GO:2001107 | negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 0.6 | 4.8 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.6 | 2.3 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.6 | 2.2 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.5 | 1.5 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.5 | 4.5 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.5 | 3.5 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.5 | 2.3 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.4 | 2.2 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.4 | 4.8 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.4 | 1.3 | GO:0032685 | negative regulation of granulocyte macrophage colony-stimulating factor production(GO:0032685) |
| 0.4 | 3.4 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.4 | 4.2 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.4 | 3.3 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.4 | 7.0 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.4 | 1.5 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.4 | 1.1 | GO:0002632 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.3 | 2.1 | GO:1901581 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.3 | 1.3 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.3 | 1.0 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.3 | 1.0 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.3 | 1.9 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) protein localization to site of double-strand break(GO:1990166) |
| 0.3 | 0.9 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.3 | 0.9 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.3 | 1.2 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.3 | 1.2 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.3 | 0.9 | GO:0086017 | Purkinje myocyte action potential(GO:0086017) Purkinje myocyte to ventricular cardiac muscle cell signaling(GO:0086029) Purkinje myocyte to ventricular cardiac muscle cell communication(GO:0086068) |
| 0.3 | 1.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.3 | 5.4 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.3 | 2.4 | GO:0015809 | arginine transport(GO:0015809) |
| 0.2 | 5.1 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.2 | 1.0 | GO:0071727 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.2 | 1.1 | GO:0060356 | leucine import(GO:0060356) |
| 0.2 | 1.3 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.2 | 0.6 | GO:1990051 | negative regulation of phospholipase C activity(GO:1900275) activation of protein kinase C activity(GO:1990051) |
| 0.2 | 1.6 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.2 | 0.9 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.2 | 1.9 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.2 | 1.0 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.2 | 1.4 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.2 | 1.3 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.2 | 0.6 | GO:0003360 | brainstem development(GO:0003360) |
| 0.1 | 1.2 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.1 | 11.2 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.1 | 1.4 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 1.0 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 | 1.6 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 0.3 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.1 | 0.7 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.7 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 5.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.5 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 1.6 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 1.4 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.1 | 2.0 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.1 | 0.4 | GO:1905171 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.1 | 4.0 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.1 | 2.6 | GO:1903077 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.1 | 1.9 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 1.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.7 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.1 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.1 | 0.4 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.7 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.0 | 1.7 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 3.5 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 3.7 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.3 | GO:0032962 | regulation of inositol trisphosphate biosynthetic process(GO:0032960) positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 1.0 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.7 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.6 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 3.5 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.0 | 0.8 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 1.1 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 1.7 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 2.0 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 1.2 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.4 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.0 | 0.4 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.3 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.9 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 1.6 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 0.7 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 0.9 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 0.8 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 3.7 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 2.3 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 1.8 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 1.5 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 3.6 | GO:0060828 | regulation of canonical Wnt signaling pathway(GO:0060828) |
| 0.0 | 0.3 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.5 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.8 | 3.9 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.5 | 2.6 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.5 | 1.4 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.3 | 2.1 | GO:1990707 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.3 | 1.2 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.3 | 1.6 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.2 | 7.0 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 1.4 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.2 | 1.6 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.2 | 0.9 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 5.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 1.3 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.2 | 0.8 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.2 | 2.0 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 4.8 | GO:0044216 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.1 | 3.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.6 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.1 | 2.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 1.0 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 4.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 3.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.0 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 2.4 | GO:0097449 | astrocyte projection(GO:0097449) postsynaptic density membrane(GO:0098839) |
| 0.1 | 1.9 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 0.4 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.7 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 2.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 1.2 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 7.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 2.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 2.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 1.6 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 1.0 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 1.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.0 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 3.5 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.9 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 4.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.7 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 2.6 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 4.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 3.4 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.9 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 1.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.9 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 2.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 2.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 4.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 8.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 21.7 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.9 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 1.0 | 2.9 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.8 | 2.4 | GO:0099530 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.7 | 7.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.6 | 2.2 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.5 | 3.5 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.5 | 1.4 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.4 | 1.3 | GO:0031726 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) CCR1 chemokine receptor binding(GO:0031726) |
| 0.4 | 1.3 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.4 | 5.1 | GO:0008061 | chitin binding(GO:0008061) |
| 0.4 | 3.3 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.4 | 6.2 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.4 | 3.8 | GO:0015288 | porin activity(GO:0015288) |
| 0.4 | 4.8 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.3 | 2.9 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.3 | 3.1 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.3 | 2.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.3 | 1.2 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.3 | 7.0 | GO:0055103 | ligase regulator activity(GO:0055103) |
| 0.3 | 5.8 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.3 | 1.3 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.3 | 1.6 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.2 | 3.5 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.2 | 4.8 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.2 | 1.6 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.2 | 2.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 12.3 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.2 | 0.9 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.2 | 1.3 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.2 | 1.0 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.2 | 1.0 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.2 | 0.9 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.2 | 6.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 1.4 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 1.5 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 0.9 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.8 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 2.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 2.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 2.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 1.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 1.0 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 4.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 1.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 2.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 2.3 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 0.2 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.1 | 0.9 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.6 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.1 | 4.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 1.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 2.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 2.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 2.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.5 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 1.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.0 | 0.7 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.2 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 1.0 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 1.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 3.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 2.9 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 1.5 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.3 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.6 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 3.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 4.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) proteasome binding(GO:0070628) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 1.0 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 9.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 2.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 7.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 1.4 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 2.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 2.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.8 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 3.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 2.4 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.7 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.3 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 1.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 2.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 18.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 3.8 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 6.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 3.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 5.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 3.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 7.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.0 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 3.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.0 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 1.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 1.0 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 1.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 3.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 3.6 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 1.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.0 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 2.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 2.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.9 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 2.3 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 2.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.7 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.8 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 1.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.1 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.4 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.0 | 0.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 2.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.7 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.4 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |