Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for T
Z-value: 1.12
Transcription factors associated with T
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
T
|
ENSMUSG00000062327.11 | T |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| T | mm39_v1_chr17_+_8653097_8653255 | -0.00 | 9.9e-01 | Click! |
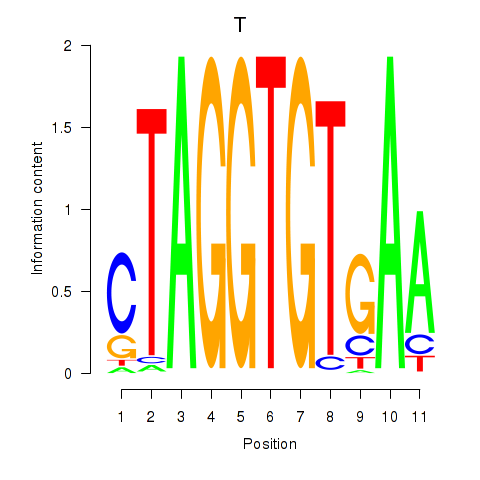
Activity profile of T motif
Sorted Z-values of T motif
Network of associatons between targets according to the STRING database.
First level regulatory network of T
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_147888816 | 12.26 |
ENSMUST00000172928.2
ENSMUST00000047315.10 |
Foxa2
|
forkhead box A2 |
| chr11_-_94283792 | 8.74 |
ENSMUST00000178136.8
ENSMUST00000021231.8 |
Abcc3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr9_-_95727267 | 8.23 |
ENSMUST00000093800.9
|
Pls1
|
plastin 1 (I-isoform) |
| chr9_+_50405817 | 7.93 |
ENSMUST00000114474.8
ENSMUST00000188047.2 |
Plet1
|
placenta expressed transcript 1 |
| chr1_+_88034556 | 7.89 |
ENSMUST00000113137.2
|
Ugt1a6b
|
UDP glucuronosyltransferase 1 family, polypeptide A6B |
| chr9_-_31375497 | 7.64 |
ENSMUST00000217007.2
ENSMUST00000213807.2 |
Tmem45b
|
transmembrane protein 45b |
| chrX_+_138511360 | 7.56 |
ENSMUST00000113026.2
|
Rnf128
|
ring finger protein 128 |
| chr3_-_107851021 | 6.14 |
ENSMUST00000106684.8
ENSMUST00000106685.9 |
Gstm6
|
glutathione S-transferase, mu 6 |
| chr10_+_127541039 | 6.07 |
ENSMUST00000079590.7
|
Myo1a
|
myosin IA |
| chr17_+_35286293 | 5.97 |
ENSMUST00000173478.2
ENSMUST00000174876.2 |
Ly6g6c
|
lymphocyte antigen 6 complex, locus G6C |
| chr15_-_76004395 | 5.81 |
ENSMUST00000239552.1
|
EPPK1
|
epiplakin 1 |
| chr7_-_79492091 | 5.57 |
ENSMUST00000049004.8
|
Anpep
|
alanyl (membrane) aminopeptidase |
| chr9_-_99592058 | 5.51 |
ENSMUST00000136429.8
|
Cldn18
|
claudin 18 |
| chr1_+_171246593 | 5.42 |
ENSMUST00000171362.2
|
Tstd1
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1 |
| chr15_-_101759212 | 5.37 |
ENSMUST00000023790.5
|
Krt1
|
keratin 1 |
| chr9_-_99592116 | 5.28 |
ENSMUST00000035048.12
|
Cldn18
|
claudin 18 |
| chr7_-_3680530 | 5.28 |
ENSMUST00000038743.15
|
Tmc4
|
transmembrane channel-like gene family 4 |
| chr9_-_78263025 | 5.26 |
ENSMUST00000125479.8
|
Gsta2
|
glutathione S-transferase, alpha 2 (Yc2) |
| chr1_+_88066086 | 5.20 |
ENSMUST00000014263.6
|
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr6_+_40619913 | 4.63 |
ENSMUST00000238599.2
|
Mgam
|
maltase-glucoamylase |
| chr4_-_129121676 | 4.62 |
ENSMUST00000106051.8
|
C77080
|
expressed sequence C77080 |
| chr8_+_46338498 | 4.57 |
ENSMUST00000034053.7
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr19_-_32080496 | 4.48 |
ENSMUST00000235213.2
ENSMUST00000236504.2 |
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr15_-_101588714 | 4.32 |
ENSMUST00000023786.7
|
Krt6b
|
keratin 6B |
| chr19_-_40260286 | 4.30 |
ENSMUST00000182432.2
|
Pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr5_+_73563418 | 4.29 |
ENSMUST00000031040.13
ENSMUST00000065543.8 |
Cwh43
|
cell wall biogenesis 43 C-terminal homolog |
| chr10_-_43934774 | 4.22 |
ENSMUST00000239010.2
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr8_+_46338557 | 4.18 |
ENSMUST00000210422.2
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr4_+_138559156 | 4.18 |
ENSMUST00000077582.14
|
Pla2g2a
|
phospholipase A2, group IIA (platelets, synovial fluid) |
| chr19_+_20579322 | 4.02 |
ENSMUST00000087638.4
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr19_-_20704896 | 3.98 |
ENSMUST00000025656.4
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7 |
| chr19_-_40260060 | 3.91 |
ENSMUST00000068439.13
|
Pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr4_-_129132963 | 3.78 |
ENSMUST00000097873.10
|
C77080
|
expressed sequence C77080 |
| chr2_+_74512638 | 3.73 |
ENSMUST00000142312.3
|
Hoxd11
|
homeobox D11 |
| chr16_+_93404719 | 3.72 |
ENSMUST00000039659.9
ENSMUST00000231762.2 |
Cbr1
|
carbonyl reductase 1 |
| chr6_-_94677118 | 3.46 |
ENSMUST00000101126.3
ENSMUST00000032105.11 |
Lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr15_-_82291372 | 3.35 |
ENSMUST00000230198.2
ENSMUST00000230248.2 ENSMUST00000072776.5 ENSMUST00000229911.2 |
Cyp2d10
|
cytochrome P450, family 2, subfamily d, polypeptide 10 |
| chr2_-_180844582 | 3.29 |
ENSMUST00000016511.6
|
Ptk6
|
PTK6 protein tyrosine kinase 6 |
| chr9_-_9239056 | 3.22 |
ENSMUST00000093893.12
|
Arhgap42
|
Rho GTPase activating protein 42 |
| chr15_-_101602734 | 3.07 |
ENSMUST00000023788.8
|
Krt6a
|
keratin 6A |
| chr8_-_34333573 | 2.65 |
ENSMUST00000183062.2
|
Rbpms
|
RNA binding protein gene with multiple splicing |
| chr12_-_113589576 | 2.62 |
ENSMUST00000103446.2
|
Ighv5-6
|
immunoglobulin heavy variable 5-6 |
| chr5_+_24633206 | 2.59 |
ENSMUST00000115049.9
|
Slc4a2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr16_-_32617783 | 2.57 |
ENSMUST00000115116.8
ENSMUST00000041123.9 |
Muc20
|
mucin 20 |
| chr1_+_132243849 | 2.53 |
ENSMUST00000072177.14
ENSMUST00000082125.6 |
Nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr1_-_71454041 | 2.51 |
ENSMUST00000087268.7
|
Abca12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr10_-_34294461 | 2.50 |
ENSMUST00000213269.2
ENSMUST00000099973.4 ENSMUST00000105512.8 ENSMUST00000047885.14 |
Nt5dc1
|
5'-nucleotidase domain containing 1 |
| chr17_+_25588241 | 2.37 |
ENSMUST00000160377.8
ENSMUST00000160485.8 |
Tpsg1
|
tryptase gamma 1 |
| chr3_+_40905066 | 2.35 |
ENSMUST00000191805.7
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr6_+_68026941 | 2.17 |
ENSMUST00000103316.2
|
Igkv9-120
|
immunoglobulin kappa chain variable 9-120 |
| chr6_-_69800923 | 2.14 |
ENSMUST00000103368.3
|
Igkv5-43
|
immunoglobulin kappa chain variable 5-43 |
| chr4_+_155648256 | 2.10 |
ENSMUST00000143840.2
ENSMUST00000146080.8 |
Nadk
|
NAD kinase |
| chrX_+_99669343 | 2.06 |
ENSMUST00000048962.4
|
Kif4
|
kinesin family member 4 |
| chr14_+_80237691 | 2.02 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr3_+_40905216 | 1.97 |
ENSMUST00000191872.6
ENSMUST00000200432.2 |
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr2_-_51862941 | 1.96 |
ENSMUST00000145481.8
ENSMUST00000112705.9 |
Nmi
|
N-myc (and STAT) interactor |
| chr12_+_119356318 | 1.76 |
ENSMUST00000221866.2
|
Macc1
|
metastasis associated in colon cancer 1 |
| chr7_-_89166781 | 1.66 |
ENSMUST00000041761.7
|
Prss23
|
protease, serine 23 |
| chr13_-_100382831 | 1.65 |
ENSMUST00000049789.3
|
Naip5
|
NLR family, apoptosis inhibitory protein 5 |
| chr15_+_95688763 | 1.62 |
ENSMUST00000227791.2
|
Ano6
|
anoctamin 6 |
| chr11_-_117670430 | 1.60 |
ENSMUST00000143406.8
|
Tmc6
|
transmembrane channel-like gene family 6 |
| chr1_-_57010921 | 1.60 |
ENSMUST00000114415.10
|
Satb2
|
special AT-rich sequence binding protein 2 |
| chr15_+_83447784 | 1.59 |
ENSMUST00000047419.8
|
Tspo
|
translocator protein |
| chr1_-_93373364 | 1.51 |
ENSMUST00000190321.7
ENSMUST00000042498.14 |
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr11_-_61233647 | 1.48 |
ENSMUST00000134423.3
ENSMUST00000093029.9 |
Slc47a2
|
solute carrier family 47, member 2 |
| chr1_-_93373145 | 1.43 |
ENSMUST00000186787.7
|
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr2_-_51863203 | 1.39 |
ENSMUST00000028314.9
|
Nmi
|
N-myc (and STAT) interactor |
| chr9_-_43151179 | 1.37 |
ENSMUST00000034512.7
|
Oaf
|
out at first homolog |
| chr5_-_38649291 | 1.33 |
ENSMUST00000129099.8
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr12_-_113561594 | 1.32 |
ENSMUST00000103444.3
|
Ighv5-4
|
immunoglobulin heavy variable 5-4 |
| chr7_-_89166724 | 1.32 |
ENSMUST00000208888.2
|
Prss23
|
protease, serine 23 |
| chr15_+_79025523 | 1.29 |
ENSMUST00000040077.8
|
Polr2f
|
polymerase (RNA) II (DNA directed) polypeptide F |
| chr12_-_113700190 | 1.27 |
ENSMUST00000103452.3
ENSMUST00000192264.2 |
Ighv5-9-1
|
immunoglobulin heavy variable 5-9-1 |
| chr19_+_37686240 | 1.23 |
ENSMUST00000025946.7
|
Cyp26a1
|
cytochrome P450, family 26, subfamily a, polypeptide 1 |
| chr11_+_78427219 | 1.21 |
ENSMUST00000050366.15
ENSMUST00000108275.2 |
Ift20
|
intraflagellar transport 20 |
| chr17_+_84819260 | 1.19 |
ENSMUST00000047206.7
|
Plekhh2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2 |
| chr14_-_75829389 | 1.16 |
ENSMUST00000165569.3
ENSMUST00000035243.5 |
Cby2
|
chibby family member 2 |
| chr9_+_121548893 | 1.14 |
ENSMUST00000182337.2
|
Nktr
|
natural killer tumor recognition sequence |
| chr4_+_53440389 | 1.13 |
ENSMUST00000107646.9
ENSMUST00000102911.10 |
Slc44a1
|
solute carrier family 44, member 1 |
| chr5_+_110434172 | 1.09 |
ENSMUST00000007296.12
ENSMUST00000112482.2 |
Pole
|
polymerase (DNA directed), epsilon |
| chr16_+_84631789 | 1.05 |
ENSMUST00000114184.8
|
Gabpa
|
GA repeat binding protein, alpha |
| chr12_-_113625906 | 1.03 |
ENSMUST00000103448.3
|
Ighv5-9
|
immunoglobulin heavy variable 5-9 |
| chr6_+_52691204 | 1.01 |
ENSMUST00000138040.8
ENSMUST00000129660.2 |
Tax1bp1
|
Tax1 (human T cell leukemia virus type I) binding protein 1 |
| chr7_-_30335277 | 1.00 |
ENSMUST00000108147.3
|
Etv2
|
ets variant 2 |
| chr6_+_139987275 | 1.00 |
ENSMUST00000043797.6
|
Capza3
|
capping protein (actin filament) muscle Z-line, alpha 3 |
| chr15_+_95688712 | 0.93 |
ENSMUST00000071874.8
|
Ano6
|
anoctamin 6 |
| chr9_-_107166543 | 0.93 |
ENSMUST00000192054.2
|
Mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr2_+_154975419 | 0.91 |
ENSMUST00000029126.15
ENSMUST00000109685.8 |
Itch
|
itchy, E3 ubiquitin protein ligase |
| chr7_-_141023199 | 0.90 |
ENSMUST00000106005.9
|
Pidd1
|
p53 induced death domain protein 1 |
| chr8_-_15096046 | 0.90 |
ENSMUST00000050493.4
ENSMUST00000123331.2 |
BB014433
|
expressed sequence BB014433 |
| chr7_+_140461860 | 0.89 |
ENSMUST00000026560.14
|
Psmd13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr7_+_140462343 | 0.89 |
ENSMUST00000163610.9
ENSMUST00000164681.8 |
Psmd13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr12_-_113823290 | 0.88 |
ENSMUST00000103459.5
|
Ighv5-17
|
immunoglobulin heavy variable 5-17 |
| chr1_-_156767196 | 0.88 |
ENSMUST00000185198.7
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr16_+_27207722 | 0.87 |
ENSMUST00000039443.14
ENSMUST00000096127.11 |
Ccdc50
|
coiled-coil domain containing 50 |
| chr10_+_23672842 | 0.87 |
ENSMUST00000119597.8
ENSMUST00000179321.8 ENSMUST00000133289.2 |
Slc18b1
|
solute carrier family 18, subfamily B, member 1 |
| chr7_-_108529375 | 0.85 |
ENSMUST00000055745.5
|
Nlrp10
|
NLR family, pyrin domain containing 10 |
| chr11_-_100628979 | 0.83 |
ENSMUST00000155500.2
ENSMUST00000107364.8 ENSMUST00000019317.12 |
Rab5c
|
RAB5C, member RAS oncogene family |
| chr7_-_71956332 | 0.82 |
ENSMUST00000079323.8
|
Mctp2
|
multiple C2 domains, transmembrane 2 |
| chr11_-_78427061 | 0.81 |
ENSMUST00000017759.9
ENSMUST00000108277.3 |
Tnfaip1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr1_-_13197387 | 0.81 |
ENSMUST00000047577.7
|
Prdm14
|
PR domain containing 14 |
| chr7_+_89779564 | 0.80 |
ENSMUST00000208742.2
ENSMUST00000049537.9 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr19_+_34078333 | 0.79 |
ENSMUST00000025685.8
|
Lipm
|
lipase, family member M |
| chr16_-_4377640 | 0.78 |
ENSMUST00000005862.9
|
Tfap4
|
transcription factor AP4 |
| chr5_+_100054110 | 0.78 |
ENSMUST00000198837.3
|
Vamp9
|
vesicle-associated membrane protein 9 |
| chr2_+_74528071 | 0.76 |
ENSMUST00000059272.10
|
Hoxd9
|
homeobox D9 |
| chr14_+_6226418 | 0.72 |
ENSMUST00000112625.9
|
Oxsm
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr9_-_106421834 | 0.72 |
ENSMUST00010181660.1
ENSMUST00000215525.2 |
ENSMUSG00001074846.1
ENSMUSG00000118396.3
|
IQ motif containing F3 IQ motif containing F3 |
| chr6_-_119444157 | 0.72 |
ENSMUST00000118120.8
|
Wnt5b
|
wingless-type MMTV integration site family, member 5B |
| chr9_-_119766580 | 0.70 |
ENSMUST00000035099.9
|
Gorasp1
|
golgi reassembly stacking protein 1 |
| chr5_-_142892457 | 0.70 |
ENSMUST00000167721.8
ENSMUST00000163829.2 ENSMUST00000100497.11 |
Actb
|
actin, beta |
| chr7_+_6200089 | 0.67 |
ENSMUST00000037553.6
|
Galp
|
galanin-like peptide |
| chr16_+_84631956 | 0.67 |
ENSMUST00000009120.8
|
Gabpa
|
GA repeat binding protein, alpha |
| chr10_+_21854540 | 0.65 |
ENSMUST00000142174.8
ENSMUST00000164659.8 |
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr13_-_21740190 | 0.64 |
ENSMUST00000068163.2
|
Olfr1535
|
olfactory receptor 1535 |
| chr10_-_23977810 | 0.64 |
ENSMUST00000170267.3
|
Taar8c
|
trace amine-associated receptor 8C |
| chr18_-_43610829 | 0.63 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr17_+_84013575 | 0.63 |
ENSMUST00000112350.8
ENSMUST00000112349.9 ENSMUST00000112352.10 ENSMUST00000067826.15 |
Mta3
|
metastasis associated 3 |
| chr10_+_128158328 | 0.63 |
ENSMUST00000219037.2
ENSMUST00000026446.4 |
Cnpy2
|
canopy FGF signaling regulator 2 |
| chr15_+_79775894 | 0.62 |
ENSMUST00000177483.8
|
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr16_-_10614679 | 0.61 |
ENSMUST00000023144.6
|
Prm1
|
protamine 1 |
| chr2_-_27138347 | 0.60 |
ENSMUST00000139312.8
|
Sardh
|
sarcosine dehydrogenase |
| chr14_-_21790463 | 0.60 |
ENSMUST00000120984.9
|
Dusp13
|
dual specificity phosphatase 13 |
| chr11_+_83553400 | 0.59 |
ENSMUST00000019074.4
|
Ccl4
|
chemokine (C-C motif) ligand 4 |
| chr18_-_36859732 | 0.59 |
ENSMUST00000061829.8
|
Cd14
|
CD14 antigen |
| chr7_-_119694400 | 0.56 |
ENSMUST00000209154.3
ENSMUST00000046993.4 |
Dnah3
|
dynein, axonemal, heavy chain 3 |
| chr7_+_43093507 | 0.55 |
ENSMUST00000004729.5
ENSMUST00000206286.2 ENSMUST00000206196.2 ENSMUST00000206411.2 |
Etfb
|
electron transferring flavoprotein, beta polypeptide |
| chr9_-_53521585 | 0.54 |
ENSMUST00000034547.6
|
Acat1
|
acetyl-Coenzyme A acetyltransferase 1 |
| chr4_-_82778066 | 0.54 |
ENSMUST00000107239.8
|
Zdhhc21
|
zinc finger, DHHC domain containing 21 |
| chr11_-_86435579 | 0.50 |
ENSMUST00000138810.3
ENSMUST00000058286.9 ENSMUST00000154617.8 |
Rps6kb1
|
ribosomal protein S6 kinase, polypeptide 1 |
| chr15_-_98155173 | 0.47 |
ENSMUST00000060855.7
|
H1f7
|
H1.7 linker histone |
| chr17_-_78991691 | 0.47 |
ENSMUST00000145480.2
|
Strn
|
striatin, calmodulin binding protein |
| chr10_-_13053733 | 0.46 |
ENSMUST00000019954.6
|
Zc2hc1b
|
zinc finger, C2HC-type containing 1B |
| chr9_-_69668204 | 0.45 |
ENSMUST00000071281.5
|
Foxb1
|
forkhead box B1 |
| chr7_-_79443536 | 0.43 |
ENSMUST00000032760.6
|
Mesp1
|
mesoderm posterior 1 |
| chr15_+_79775819 | 0.42 |
ENSMUST00000177350.8
|
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr15_-_60793115 | 0.41 |
ENSMUST00000096418.5
|
A1bg
|
alpha-1-B glycoprotein |
| chr2_-_125993887 | 0.41 |
ENSMUST00000110448.3
ENSMUST00000110446.9 |
Fam227b
|
family with sequence similarity 227, member B |
| chr11_+_78427181 | 0.40 |
ENSMUST00000128788.8
|
Ift20
|
intraflagellar transport 20 |
| chr5_-_151510389 | 0.39 |
ENSMUST00000165928.4
|
Vmn2r18
|
vomeronasal 2, receptor 18 |
| chr2_+_181134977 | 0.37 |
ENSMUST00000069649.9
|
Abhd16b
|
abhydrolase domain containing 16B |
| chr14_+_13803477 | 0.36 |
ENSMUST00000102996.4
|
Cfap20dc
|
CFAP20 domain containing |
| chr8_-_73246435 | 0.36 |
ENSMUST00000152080.8
|
Slc35e1
|
solute carrier family 35, member E1 |
| chr4_+_52964547 | 0.35 |
ENSMUST00000215010.2
ENSMUST00000215127.2 |
Olfr270
|
olfactory receptor 270 |
| chr5_-_108280350 | 0.35 |
ENSMUST00000119784.2
ENSMUST00000117759.2 |
Tmed5
|
transmembrane p24 trafficking protein 5 |
| chr9_+_37524966 | 0.33 |
ENSMUST00000215474.2
|
Siae
|
sialic acid acetylesterase |
| chr12_-_111933328 | 0.33 |
ENSMUST00000222874.2
ENSMUST00000223191.2 ENSMUST00000021719.7 |
Atp5mpl
|
ATP synthase membrane subunit 6.8PL |
| chr15_+_9335636 | 0.32 |
ENSMUST00000072403.7
|
Ugt3a2
|
UDP glycosyltransferases 3 family, polypeptide A2 |
| chr1_+_44158111 | 0.29 |
ENSMUST00000155917.8
|
Bivm
|
basic, immunoglobulin-like variable motif containing |
| chr11_+_29476385 | 0.27 |
ENSMUST00000133452.8
|
Mtif2
|
mitochondrial translational initiation factor 2 |
| chr8_+_114369838 | 0.26 |
ENSMUST00000095173.3
ENSMUST00000034219.12 ENSMUST00000212269.2 |
Syce1l
|
synaptonemal complex central element protein 1 like |
| chr10_+_88295515 | 0.23 |
ENSMUST00000125612.2
|
Sycp3
|
synaptonemal complex protein 3 |
| chr14_+_54434705 | 0.22 |
ENSMUST00000103719.2
|
Traj22
|
T cell receptor alpha joining 22 |
| chr12_-_113666198 | 0.22 |
ENSMUST00000103450.4
|
Ighv5-12
|
immunoglobulin heavy variable 5-12 |
| chr1_-_86317066 | 0.21 |
ENSMUST00000212058.2
|
Nmur1
|
neuromedin U receptor 1 |
| chr8_+_121262528 | 0.21 |
ENSMUST00000120493.8
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chr7_+_15246265 | 0.20 |
ENSMUST00000172881.8
|
Obox1
|
oocyte specific homeobox 1 |
| chr4_+_146533480 | 0.19 |
ENSMUST00000105733.3
|
Zfp992
|
zinc finger protein 992 |
| chr8_+_3520841 | 0.19 |
ENSMUST00000047265.13
|
Tex45
|
testis expressed 45 |
| chr13_-_113317431 | 0.17 |
ENSMUST00000038212.14
|
Gzmk
|
granzyme K |
| chr8_-_83129832 | 0.16 |
ENSMUST00000034148.7
|
Il15
|
interleukin 15 |
| chr6_+_90310252 | 0.16 |
ENSMUST00000046128.12
ENSMUST00000164761.6 |
Uroc1
|
urocanase domain containing 1 |
| chr11_+_87938626 | 0.14 |
ENSMUST00000107920.10
|
Srsf1
|
serine and arginine-rich splicing factor 1 |
| chr2_+_129965195 | 0.13 |
ENSMUST00000028888.5
|
Tgm6
|
transglutaminase 6 |
| chr15_+_79776597 | 0.12 |
ENSMUST00000175714.8
ENSMUST00000109620.10 ENSMUST00000165537.8 ENSMUST00000175752.8 ENSMUST00000176325.8 |
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr7_-_140462221 | 0.12 |
ENSMUST00000026559.14
|
Sirt3
|
sirtuin 3 |
| chr2_+_164790139 | 0.11 |
ENSMUST00000017881.3
|
Mmp9
|
matrix metallopeptidase 9 |
| chr16_-_88504848 | 0.11 |
ENSMUST00000050882.4
|
2310061N02Rik
|
RIKEN cDNA 2310061N02 gene |
| chr3_+_90138895 | 0.10 |
ENSMUST00000029546.15
ENSMUST00000119304.2 |
Jtb
|
jumping translocation breakpoint |
| chr3_-_103698391 | 0.10 |
ENSMUST00000106845.9
ENSMUST00000029438.15 |
Hipk1
|
homeodomain interacting protein kinase 1 |
| chr7_-_130931235 | 0.09 |
ENSMUST00000188899.2
|
Fam24b
|
family with sequence similarity 24 member B |
| chr5_+_129924619 | 0.09 |
ENSMUST00000077320.3
|
Zbed5
|
zinc finger, BED type containing 5 |
| chr11_+_79551358 | 0.08 |
ENSMUST00000155381.2
|
Rab11fip4
|
RAB11 family interacting protein 4 (class II) |
| chr6_-_139987135 | 0.07 |
ENSMUST00000032356.13
|
Plcz1
|
phospholipase C, zeta 1 |
| chr6_+_72281587 | 0.06 |
ENSMUST00000183018.8
ENSMUST00000182014.10 |
Sftpb
|
surfactant associated protein B |
| chr5_+_129924564 | 0.06 |
ENSMUST00000041466.14
|
Zbed5
|
zinc finger, BED type containing 5 |
| chrX_-_142716085 | 0.03 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr8_+_61085853 | 0.03 |
ENSMUST00000161421.2
|
Mfap3l
|
microfibrillar-associated protein 3-like |
| chr15_-_79025387 | 0.01 |
ENSMUST00000187550.7
ENSMUST00000188562.7 ENSMUST00000190509.7 ENSMUST00000190730.7 ENSMUST00000190959.7 ENSMUST00000169604.8 ENSMUST00000186053.7 |
1700088E04Rik
|
RIKEN cDNA 1700088E04 gene |
| chr7_-_15361801 | 0.01 |
ENSMUST00000086122.10
ENSMUST00000174443.8 |
Obox3
|
oocyte specific homeobox 3 |
| chrX_-_110446022 | 0.00 |
ENSMUST00000156639.2
|
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.3 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 2.1 | 8.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 1.5 | 10.8 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 1.5 | 8.7 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 1.2 | 3.5 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 1.1 | 3.4 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 1.1 | 13.1 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.9 | 2.6 | GO:0002543 | activation of blood coagulation via clotting cascade(GO:0002543) phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.8 | 2.5 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.6 | 3.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.5 | 3.7 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.4 | 4.0 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.4 | 5.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.4 | 1.1 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.3 | 5.2 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.3 | 5.3 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.2 | 2.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.2 | 0.7 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.2 | 1.6 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.2 | 3.7 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.2 | 7.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.2 | 0.4 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.2 | 7.9 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) |
| 0.2 | 0.8 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.2 | 0.6 | GO:1901053 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.2 | 1.0 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.2 | 0.6 | GO:0071726 | response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.2 | 6.1 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.2 | 1.7 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.2 | 2.1 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.2 | 0.9 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.2 | 0.5 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.2 | 7.6 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.2 | 5.3 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.2 | 0.3 | GO:0080184 | response to phenylpropanoid(GO:0080184) |
| 0.1 | 0.6 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 1.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 3.3 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 0.8 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 1.6 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.1 | 2.6 | GO:0048012 | hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.1 | 2.6 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.1 | 1.6 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.5 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.1 | 1.8 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.8 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.1 | 0.8 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 0.7 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.1 | 1.5 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.1 | 0.6 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.1 | 0.9 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.9 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 0.5 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 | 4.3 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 1.3 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 0.8 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.1 | 0.9 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 1.2 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.1 | 0.9 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 7.2 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.5 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.6 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.8 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 2.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 2.0 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.1 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 1.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.6 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.2 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 0.0 | 2.2 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 0.8 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 8.7 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.2 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) extrathymic T cell selection(GO:0045062) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.7 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.3 | GO:0007128 | meiotic prophase I(GO:0007128) prophase(GO:0051324) |
| 0.0 | 0.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 2.6 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.1 | GO:0051878 | lateral element assembly(GO:0051878) |
| 0.0 | 0.6 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 | 0.4 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 4.3 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 3.2 | GO:0006665 | sphingolipid metabolic process(GO:0006665) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 2.5 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.6 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 1.0 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 8.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.5 | 1.6 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.5 | 2.5 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.4 | 5.6 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.2 | 1.6 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.2 | 1.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.2 | 8.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.8 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.5 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.1 | 0.8 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 1.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 2.6 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 8.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 2.0 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 0.6 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 2.9 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.0 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 7.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 11.3 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 7.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.6 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 9.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 10.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 7.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 2.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 3.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 5.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 2.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.7 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 2.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 4.1 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 0.1 | GO:0000802 | transverse filament(GO:0000802) |
| 0.0 | 1.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 40.0 | GO:0070062 | extracellular exosome(GO:0070062) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 8.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 1.0 | 8.7 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.9 | 5.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.8 | 2.5 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.8 | 4.6 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.6 | 4.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.5 | 3.7 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.5 | 2.6 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.3 | 13.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.3 | 1.6 | GO:0002046 | opsin binding(GO:0002046) |
| 0.3 | 8.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.2 | 0.7 | GO:0098918 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.2 | 5.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 1.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.2 | 0.6 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.2 | 11.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 0.6 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.2 | 1.2 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.2 | 1.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.2 | 12.3 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.7 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.1 | 8.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.5 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 2.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 2.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 1.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 1.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) RNA polymerase II activity(GO:0001055) |
| 0.1 | 3.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.6 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 0.6 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 1.3 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.5 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.1 | 2.1 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 0.2 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.1 | 2.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 1.5 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.1 | 0.6 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.9 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 7.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.8 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 14.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.8 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 1.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 1.1 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 14.4 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 4.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.7 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 5.0 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.6 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.2 | 12.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 7.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 3.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 5.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 5.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.5 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.7 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.4 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.6 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 2.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.8 | 8.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.4 | 4.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.4 | 12.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.3 | 10.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 4.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 5.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 2.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 4.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 1.1 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.6 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.9 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 1.2 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.8 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 1.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.5 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 3.5 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 1.0 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.7 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 2.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |