Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Tal1
Z-value: 1.96
Transcription factors associated with Tal1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tal1
|
ENSMUSG00000028717.13 | Tal1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tal1 | mm39_v1_chr4_+_114916703_114916759 | 0.81 | 4.0e-18 | Click! |
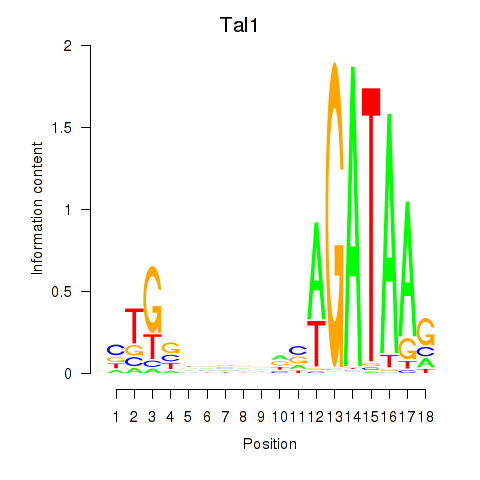
Activity profile of Tal1 motif
Sorted Z-values of Tal1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tal1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_103159892 | 29.64 |
ENSMUST00000133600.8
ENSMUST00000134554.2 ENSMUST00000156927.8 |
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr15_-_103160082 | 24.72 |
ENSMUST00000149111.8
ENSMUST00000132836.8 |
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr4_+_132701413 | 21.84 |
ENSMUST00000030693.13
|
Fgr
|
FGR proto-oncogene, Src family tyrosine kinase |
| chr17_+_41121979 | 21.08 |
ENSMUST00000024721.8
ENSMUST00000233740.2 |
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr17_+_37180437 | 18.84 |
ENSMUST00000060524.11
|
Trim10
|
tripartite motif-containing 10 |
| chr1_+_131566044 | 16.79 |
ENSMUST00000073350.13
|
Ctse
|
cathepsin E |
| chr8_+_81220410 | 16.42 |
ENSMUST00000063359.8
|
Gypa
|
glycophorin A |
| chr1_+_131566223 | 16.27 |
ENSMUST00000112411.2
|
Ctse
|
cathepsin E |
| chr1_+_174000304 | 15.73 |
ENSMUST00000027817.8
|
Spta1
|
spectrin alpha, erythrocytic 1 |
| chr5_+_90920294 | 15.55 |
ENSMUST00000031320.8
|
Pf4
|
platelet factor 4 |
| chr5_+_90920353 | 14.88 |
ENSMUST00000202625.2
|
Pf4
|
platelet factor 4 |
| chr3_-_14873406 | 14.38 |
ENSMUST00000181860.8
ENSMUST00000144327.3 |
Car1
|
carbonic anhydrase 1 |
| chr6_+_67586695 | 13.37 |
ENSMUST00000103303.3
|
Igkv1-135
|
immunoglobulin kappa variable 1-135 |
| chr4_-_119047202 | 13.23 |
ENSMUST00000239029.2
ENSMUST00000138395.9 ENSMUST00000156746.3 |
Ermap
|
erythroblast membrane-associated protein |
| chr4_+_134591847 | 12.94 |
ENSMUST00000030627.8
|
Rhd
|
Rh blood group, D antigen |
| chr7_-_4400704 | 12.60 |
ENSMUST00000108590.4
ENSMUST00000206928.2 |
Gp6
|
glycoprotein 6 (platelet) |
| chr8_+_85428059 | 12.56 |
ENSMUST00000238364.2
ENSMUST00000238562.2 ENSMUST00000037165.6 |
Lyl1
|
lymphoblastomic leukemia 1 |
| chr14_-_44112974 | 12.53 |
ENSMUST00000179200.2
|
Ear1
|
eosinophil-associated, ribonuclease A family, member 1 |
| chr14_-_44057096 | 12.15 |
ENSMUST00000100691.4
|
Ear1
|
eosinophil-associated, ribonuclease A family, member 1 |
| chr19_+_58658779 | 12.08 |
ENSMUST00000057270.9
|
Pnlip
|
pancreatic lipase |
| chr4_-_119047167 | 11.45 |
ENSMUST00000030396.15
|
Ermap
|
erythroblast membrane-associated protein |
| chr14_+_44340111 | 11.04 |
ENSMUST00000074839.7
|
Ear2
|
eosinophil-associated, ribonuclease A family, member 2 |
| chr15_-_74635423 | 10.97 |
ENSMUST00000040404.8
|
Ly6d
|
lymphocyte antigen 6 complex, locus D |
| chr15_+_80507671 | 10.96 |
ENSMUST00000043149.9
|
Grap2
|
GRB2-related adaptor protein 2 |
| chrX_+_8137881 | 10.66 |
ENSMUST00000115590.2
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr19_+_58658838 | 9.71 |
ENSMUST00000238108.2
|
Pnlip
|
pancreatic lipase |
| chr6_+_41279199 | 9.23 |
ENSMUST00000031913.5
|
Try4
|
trypsin 4 |
| chr14_+_51366512 | 9.22 |
ENSMUST00000095923.4
|
Rnase6
|
ribonuclease, RNase A family, 6 |
| chr17_-_26417982 | 9.09 |
ENSMUST00000142410.2
ENSMUST00000120333.8 ENSMUST00000039113.14 |
Pdia2
|
protein disulfide isomerase associated 2 |
| chr9_-_44473146 | 8.97 |
ENSMUST00000215293.2
|
Cxcr5
|
chemokine (C-X-C motif) receptor 5 |
| chr9_-_32452885 | 8.96 |
ENSMUST00000016231.14
|
Fli1
|
Friend leukemia integration 1 |
| chr1_-_132295617 | 8.77 |
ENSMUST00000142609.8
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr14_+_52091156 | 8.57 |
ENSMUST00000169070.2
ENSMUST00000074477.7 |
Ear6
|
eosinophil-associated, ribonuclease A family, member 6 |
| chr1_-_181669891 | 8.52 |
ENSMUST00000193028.2
ENSMUST00000191878.6 ENSMUST00000005003.12 |
Lbr
|
lamin B receptor |
| chr9_-_21874802 | 8.42 |
ENSMUST00000006397.7
|
Epor
|
erythropoietin receptor |
| chr4_-_119047180 | 8.17 |
ENSMUST00000150864.3
ENSMUST00000141227.9 |
Ermap
|
erythroblast membrane-associated protein |
| chr11_-_79971750 | 8.11 |
ENSMUST00000103233.10
ENSMUST00000061283.15 |
Crlf3
|
cytokine receptor-like factor 3 |
| chr4_-_119047146 | 7.89 |
ENSMUST00000124626.9
|
Ermap
|
erythroblast membrane-associated protein |
| chr2_+_84669739 | 7.85 |
ENSMUST00000146816.8
ENSMUST00000028469.14 |
Slc43a1
|
solute carrier family 43, member 1 |
| chr2_-_102731691 | 7.34 |
ENSMUST00000111192.3
ENSMUST00000111190.9 ENSMUST00000111198.9 ENSMUST00000111191.9 ENSMUST00000060516.14 ENSMUST00000099673.9 ENSMUST00000005218.15 ENSMUST00000111194.8 |
Cd44
|
CD44 antigen |
| chr11_+_58808830 | 7.21 |
ENSMUST00000020792.12
ENSMUST00000108818.4 |
Btnl10
|
butyrophilin-like 10 |
| chr8_-_86091946 | 7.12 |
ENSMUST00000034133.14
|
Mylk3
|
myosin light chain kinase 3 |
| chr1_+_165596961 | 7.10 |
ENSMUST00000040298.5
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr18_+_89195089 | 6.90 |
ENSMUST00000236644.2
ENSMUST00000236828.2 |
Cd226
|
CD226 antigen |
| chr11_-_102360664 | 6.81 |
ENSMUST00000103086.4
|
Itga2b
|
integrin alpha 2b |
| chr16_-_19801781 | 6.64 |
ENSMUST00000058839.10
|
Klhl6
|
kelch-like 6 |
| chr6_+_48624295 | 6.51 |
ENSMUST00000078223.6
ENSMUST00000203509.2 |
Gimap8
|
GTPase, IMAP family member 8 |
| chr1_-_173363523 | 6.34 |
ENSMUST00000139092.8
|
Ifi214
|
interferon activated gene 214 |
| chr12_-_115884332 | 6.09 |
ENSMUST00000103548.3
|
Ighv1-81
|
immunoglobulin heavy variable 1-81 |
| chr19_-_11058452 | 6.05 |
ENSMUST00000025636.8
|
Ms4a8a
|
membrane-spanning 4-domains, subfamily A, member 8A |
| chr9_+_20940669 | 5.99 |
ENSMUST00000001040.7
ENSMUST00000215077.2 |
Icam4
|
intercellular adhesion molecule 4, Landsteiner-Wiener blood group |
| chr1_-_171476495 | 5.79 |
ENSMUST00000194791.2
ENSMUST00000192024.6 |
Slamf7
|
SLAM family member 7 |
| chr6_+_41331039 | 5.69 |
ENSMUST00000072103.7
|
Try10
|
trypsin 10 |
| chr1_-_171476559 | 5.68 |
ENSMUST00000111276.9
ENSMUST00000194531.6 |
Slamf7
|
SLAM family member 7 |
| chr11_+_117673107 | 5.57 |
ENSMUST00000050874.14
ENSMUST00000106334.9 |
Tmc8
|
transmembrane channel-like gene family 8 |
| chr14_+_51366306 | 5.38 |
ENSMUST00000226210.2
|
Rnase6
|
ribonuclease, RNase A family, 6 |
| chr6_-_131224305 | 5.28 |
ENSMUST00000032306.15
ENSMUST00000088867.7 |
Klra2
|
killer cell lectin-like receptor, subfamily A, member 2 |
| chr6_+_48624158 | 5.22 |
ENSMUST00000203083.3
|
Gimap8
|
GTPase, IMAP family member 8 |
| chr6_-_136834725 | 5.22 |
ENSMUST00000032341.3
|
Art4
|
ADP-ribosyltransferase 4 |
| chr6_+_68518603 | 4.98 |
ENSMUST00000168090.3
ENSMUST00000103326.3 |
Igkv1-99
|
immunoglobulin kappa variable 1-99 |
| chr8_+_85428391 | 4.95 |
ENSMUST00000238338.2
|
Lyl1
|
lymphoblastomic leukemia 1 |
| chr16_-_18440388 | 4.92 |
ENSMUST00000167388.3
|
Gp1bb
|
glycoprotein Ib, beta polypeptide |
| chr9_-_110886306 | 4.92 |
ENSMUST00000195968.2
ENSMUST00000111888.3 |
Ccrl2
|
chemokine (C-C motif) receptor-like 2 |
| chr9_+_32547529 | 4.92 |
ENSMUST00000238819.2
|
Ets1
|
E26 avian leukemia oncogene 1, 5' domain |
| chr1_+_171386752 | 4.89 |
ENSMUST00000004829.13
|
Cd244a
|
CD244 molecule A |
| chr1_+_165591315 | 4.80 |
ENSMUST00000111432.10
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr3_-_83749036 | 4.79 |
ENSMUST00000029623.11
|
Tlr2
|
toll-like receptor 2 |
| chr11_+_94827050 | 4.72 |
ENSMUST00000001547.8
|
Col1a1
|
collagen, type I, alpha 1 |
| chr9_-_99599312 | 4.68 |
ENSMUST00000112882.9
ENSMUST00000131922.2 |
Cldn18
|
claudin 18 |
| chr12_-_114477427 | 4.64 |
ENSMUST00000191803.2
|
Ighv1-5
|
immunoglobulin heavy variable V1-5 |
| chr19_+_58748132 | 4.59 |
ENSMUST00000026081.5
|
Pnliprp2
|
pancreatic lipase-related protein 2 |
| chr6_+_30541581 | 4.40 |
ENSMUST00000096066.5
|
Cpa2
|
carboxypeptidase A2, pancreatic |
| chr16_+_58490625 | 4.22 |
ENSMUST00000060077.7
|
Cpox
|
coproporphyrinogen oxidase |
| chr11_+_117673198 | 4.21 |
ENSMUST00000117781.8
|
Tmc8
|
transmembrane channel-like gene family 8 |
| chr14_+_26722319 | 4.16 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr8_-_71964379 | 4.09 |
ENSMUST00000048452.6
|
Plvap
|
plasmalemma vesicle associated protein |
| chr15_+_78810919 | 3.98 |
ENSMUST00000089377.6
|
Lgals1
|
lectin, galactose binding, soluble 1 |
| chr14_-_44161016 | 3.92 |
ENSMUST00000159175.2
|
Ear10
|
eosinophil-associated, ribonuclease A family, member 10 |
| chr17_-_33932346 | 3.88 |
ENSMUST00000173392.2
|
Marchf2
|
membrane associated ring-CH-type finger 2 |
| chr4_-_155012643 | 3.87 |
ENSMUST00000123514.8
|
Tnfrsf14
|
tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) |
| chr5_-_53864595 | 3.87 |
ENSMUST00000200691.4
|
Cckar
|
cholecystokinin A receptor |
| chr9_-_110886576 | 3.84 |
ENSMUST00000199839.5
|
Ccrl2
|
chemokine (C-C motif) receptor-like 2 |
| chr16_+_22965286 | 3.80 |
ENSMUST00000023593.6
|
Adipoq
|
adiponectin, C1Q and collagen domain containing |
| chr4_-_59438633 | 3.79 |
ENSMUST00000040166.14
ENSMUST00000107544.2 |
Susd1
|
sushi domain containing 1 |
| chr12_-_114528632 | 3.76 |
ENSMUST00000195337.2
ENSMUST00000103497.2 |
Ighv15-2
|
immunoglobulin heavy variable V15-2 |
| chr7_+_4467730 | 3.73 |
ENSMUST00000086372.8
ENSMUST00000169820.8 ENSMUST00000163893.8 ENSMUST00000170635.2 |
Eps8l1
|
EPS8-like 1 |
| chr5_+_137628377 | 3.64 |
ENSMUST00000175968.8
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr6_+_17306334 | 3.62 |
ENSMUST00000007799.13
ENSMUST00000115456.6 |
Cav1
|
caveolin 1, caveolae protein |
| chr6_+_17306414 | 3.61 |
ENSMUST00000150901.2
|
Cav1
|
caveolin 1, caveolae protein |
| chr5_-_105258142 | 3.61 |
ENSMUST00000031238.13
|
Gbp9
|
guanylate-binding protein 9 |
| chr14_+_22069877 | 3.51 |
ENSMUST00000161249.8
ENSMUST00000159777.8 ENSMUST00000162540.2 |
Lrmda
|
leucine rich melanocyte differentiation associated |
| chr5_+_115604321 | 3.48 |
ENSMUST00000145785.8
ENSMUST00000031495.11 ENSMUST00000112071.8 ENSMUST00000125568.2 |
Pla2g1b
|
phospholipase A2, group IB, pancreas |
| chr8_+_23525101 | 3.41 |
ENSMUST00000117662.8
ENSMUST00000117296.8 ENSMUST00000141784.9 |
Ank1
|
ankyrin 1, erythroid |
| chr6_+_17306379 | 3.41 |
ENSMUST00000115455.3
|
Cav1
|
caveolin 1, caveolae protein |
| chrX_+_55493325 | 3.32 |
ENSMUST00000079663.7
|
Gm2174
|
predicted gene 2174 |
| chr5_-_105287405 | 3.31 |
ENSMUST00000100961.5
ENSMUST00000031235.13 ENSMUST00000197799.2 ENSMUST00000199629.2 ENSMUST00000196677.5 ENSMUST00000100962.8 |
Gbp9
Gbp8
Gbp4
|
guanylate-binding protein 9 guanylate-binding protein 8 guanylate binding protein 4 |
| chr6_-_68732577 | 3.25 |
ENSMUST00000103332.2
|
Igkv4-92
|
immunoglobulin kappa variable 4-92 |
| chr16_+_49620883 | 3.25 |
ENSMUST00000229640.2
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr6_-_87567690 | 3.14 |
ENSMUST00000203636.2
ENSMUST00000050887.9 ENSMUST00000204682.3 |
Prokr1
|
prokineticin receptor 1 |
| chr6_+_86055018 | 3.13 |
ENSMUST00000205034.3
ENSMUST00000203724.3 |
Add2
|
adducin 2 (beta) |
| chr9_+_59198829 | 3.12 |
ENSMUST00000217570.2
ENSMUST00000026266.9 |
Adpgk
|
ADP-dependent glucokinase |
| chr2_+_85850977 | 3.08 |
ENSMUST00000213774.2
ENSMUST00000214546.2 ENSMUST00000215682.2 |
Olfr1033
|
olfactory receptor 1033 |
| chr17_+_31427023 | 3.08 |
ENSMUST00000173776.2
ENSMUST00000048656.15 |
Ubash3a
|
ubiquitin associated and SH3 domain containing, A |
| chr6_+_67701864 | 3.07 |
ENSMUST00000103304.3
|
Igkv1-133
|
immunoglobulin kappa variable 1-133 |
| chr3_+_79791798 | 2.98 |
ENSMUST00000118853.8
ENSMUST00000145992.2 |
Gask1b
|
golgi associated kinase 1B |
| chr18_-_36859732 | 2.88 |
ENSMUST00000061829.8
|
Cd14
|
CD14 antigen |
| chr10_-_117060377 | 2.87 |
ENSMUST00000020382.8
|
Yeats4
|
YEATS domain containing 4 |
| chr11_+_117672902 | 2.83 |
ENSMUST00000127080.9
|
Tmc8
|
transmembrane channel-like gene family 8 |
| chr2_-_75534985 | 2.76 |
ENSMUST00000102672.5
|
Nfe2l2
|
nuclear factor, erythroid derived 2, like 2 |
| chr12_-_113928438 | 2.67 |
ENSMUST00000103478.4
|
Ighv3-1
|
immunoglobulin heavy variable 3-1 |
| chr2_-_66615247 | 2.67 |
ENSMUST00000042792.7
|
Scn7a
|
sodium channel, voltage-gated, type VII, alpha |
| chr2_-_153083322 | 2.65 |
ENSMUST00000056924.14
|
Plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr5_-_24235295 | 2.63 |
ENSMUST00000101513.9
|
Fam126a
|
family with sequence similarity 126, member A |
| chr13_-_95661726 | 2.63 |
ENSMUST00000022185.10
|
F2rl1
|
coagulation factor II (thrombin) receptor-like 1 |
| chr4_-_41464816 | 2.60 |
ENSMUST00000108055.9
ENSMUST00000154535.8 ENSMUST00000030148.6 |
Kif24
|
kinesin family member 24 |
| chr3_+_89043440 | 2.56 |
ENSMUST00000047111.13
|
Pklr
|
pyruvate kinase liver and red blood cell |
| chr16_-_18882012 | 2.49 |
ENSMUST00000199490.2
|
Iglj1
|
immunoglobulin lambda joining 1 |
| chr10_-_93727003 | 2.48 |
ENSMUST00000180840.8
|
Metap2
|
methionine aminopeptidase 2 |
| chr6_-_25809209 | 2.47 |
ENSMUST00000115330.8
|
Pot1a
|
protection of telomeres 1A |
| chr6_+_41435846 | 2.43 |
ENSMUST00000031910.8
|
Prss1
|
protease, serine 1 (trypsin 1) |
| chr7_+_99243812 | 2.40 |
ENSMUST00000162290.2
|
Arrb1
|
arrestin, beta 1 |
| chr7_+_110372860 | 2.38 |
ENSMUST00000143786.2
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr5_-_24235646 | 2.36 |
ENSMUST00000197617.5
ENSMUST00000030849.13 |
Fam126a
|
family with sequence similarity 126, member A |
| chr13_-_13568106 | 2.18 |
ENSMUST00000021738.10
ENSMUST00000220628.2 |
Gpr137b
|
G protein-coupled receptor 137B |
| chr1_-_131161312 | 2.15 |
ENSMUST00000212202.2
|
Rassf5
|
Ras association (RalGDS/AF-6) domain family member 5 |
| chr2_-_156848923 | 2.11 |
ENSMUST00000146413.8
ENSMUST00000103129.9 ENSMUST00000103130.8 |
Dsn1
|
DSN1 homolog, MIS12 kinetochore complex component |
| chr5_-_121974913 | 2.09 |
ENSMUST00000040308.14
ENSMUST00000086310.8 |
Sh2b3
|
SH2B adaptor protein 3 |
| chr5_+_123482496 | 2.08 |
ENSMUST00000031391.9
ENSMUST00000117971.2 |
Bcl7a
|
B cell CLL/lymphoma 7A |
| chr7_-_126497421 | 2.04 |
ENSMUST00000121532.8
ENSMUST00000032926.12 |
Tmem219
|
transmembrane protein 219 |
| chr10_-_128462616 | 2.03 |
ENSMUST00000026420.7
|
Rps26
|
ribosomal protein S26 |
| chr1_-_23961379 | 2.01 |
ENSMUST00000027339.14
|
Smap1
|
small ArfGAP 1 |
| chr17_+_48037758 | 1.96 |
ENSMUST00000024782.12
ENSMUST00000144955.2 |
Pgc
|
progastricsin (pepsinogen C) |
| chr16_-_13548833 | 1.96 |
ENSMUST00000023364.7
|
Pla2g10
|
phospholipase A2, group X |
| chr13_-_74882328 | 1.94 |
ENSMUST00000223309.2
|
Cast
|
calpastatin |
| chr9_-_21042521 | 1.87 |
ENSMUST00000216874.2
ENSMUST00000214454.2 ENSMUST00000214864.2 |
Tyk2
|
tyrosine kinase 2 |
| chr14_+_22069709 | 1.82 |
ENSMUST00000075639.11
|
Lrmda
|
leucine rich melanocyte differentiation associated |
| chr8_+_105066924 | 1.80 |
ENSMUST00000212081.2
|
Cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr2_-_153067297 | 1.79 |
ENSMUST00000099194.4
|
Tspyl3
|
TSPY-like 3 |
| chr11_+_50741512 | 1.79 |
ENSMUST00000171427.8
|
Grm6
|
glutamate receptor, metabotropic 6 |
| chr8_+_105066980 | 1.78 |
ENSMUST00000211885.2
|
Cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr3_+_89043879 | 1.78 |
ENSMUST00000107482.10
ENSMUST00000127058.2 |
Pklr
|
pyruvate kinase liver and red blood cell |
| chr7_-_4527228 | 1.77 |
ENSMUST00000154913.2
|
Tnni3
|
troponin I, cardiac 3 |
| chr16_+_22965330 | 1.74 |
ENSMUST00000171309.2
|
Adipoq
|
adiponectin, C1Q and collagen domain containing |
| chr11_-_117673008 | 1.74 |
ENSMUST00000152304.3
|
Tmc6
|
transmembrane channel-like gene family 6 |
| chr6_-_130363837 | 1.67 |
ENSMUST00000032288.6
|
Klra1
|
killer cell lectin-like receptor, subfamily A, member 1 |
| chr4_-_49681954 | 1.61 |
ENSMUST00000029991.3
|
Ppp3r2
|
protein phosphatase 3, regulatory subunit B, alpha isoform (calcineurin B, type II) |
| chr6_+_125048230 | 1.60 |
ENSMUST00000140346.9
ENSMUST00000171989.3 |
Lpar5
|
lysophosphatidic acid receptor 5 |
| chr8_-_71990085 | 1.60 |
ENSMUST00000051672.9
|
Bst2
|
bone marrow stromal cell antigen 2 |
| chr12_-_72711509 | 1.59 |
ENSMUST00000221750.2
|
Dhrs7
|
dehydrogenase/reductase (SDR family) member 7 |
| chr8_-_21392510 | 1.52 |
ENSMUST00000122025.9
|
Gm15056
|
predicted gene 15056 |
| chr17_-_35265702 | 1.50 |
ENSMUST00000097338.11
|
Msh5
|
mutS homolog 5 |
| chr6_+_86055048 | 1.46 |
ENSMUST00000032069.8
|
Add2
|
adducin 2 (beta) |
| chr14_+_50595361 | 1.40 |
ENSMUST00000185091.2
|
Tlr11
|
toll-like receptor 11 |
| chr11_-_78067446 | 1.38 |
ENSMUST00000148154.3
ENSMUST00000017549.13 |
Nek8
|
NIMA (never in mitosis gene a)-related expressed kinase 8 |
| chr15_-_34356567 | 1.35 |
ENSMUST00000179647.2
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr17_-_48739874 | 1.34 |
ENSMUST00000046549.5
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2 |
| chrX_+_37405054 | 1.31 |
ENSMUST00000016471.9
ENSMUST00000115134.2 |
Atp1b4
|
ATPase, (Na+)/K+ transporting, beta 4 polypeptide |
| chr5_-_137145030 | 1.29 |
ENSMUST00000054384.6
ENSMUST00000152207.2 |
Trim56
|
tripartite motif-containing 56 |
| chr6_-_34294377 | 1.25 |
ENSMUST00000154655.2
ENSMUST00000102980.11 |
Akr1b3
|
aldo-keto reductase family 1, member B3 (aldose reductase) |
| chr2_-_87504008 | 1.25 |
ENSMUST00000213835.2
|
Olfr1135
|
olfactory receptor 1135 |
| chr4_+_56802338 | 1.21 |
ENSMUST00000045368.12
|
Abitram
|
actin binding transcription modulator |
| chrX_+_21581135 | 1.19 |
ENSMUST00000033414.8
|
Slc6a14
|
solute carrier family 6 (neurotransmitter transporter), member 14 |
| chr7_-_83444026 | 1.17 |
ENSMUST00000119134.8
|
Cfap161
|
cilia and flagella associated protein 161 |
| chr5_+_20112771 | 1.15 |
ENSMUST00000200443.2
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr8_+_108162985 | 1.14 |
ENSMUST00000166615.3
ENSMUST00000213097.2 ENSMUST00000212205.2 |
Wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr2_+_88505972 | 1.12 |
ENSMUST00000216767.2
ENSMUST00000213893.2 |
Olfr1193
|
olfactory receptor 1193 |
| chr11_+_67090878 | 1.06 |
ENSMUST00000124516.8
ENSMUST00000018637.15 ENSMUST00000129018.8 |
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chr7_-_115445315 | 1.05 |
ENSMUST00000166207.3
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr6_-_38331482 | 1.04 |
ENSMUST00000031850.10
ENSMUST00000114898.3 |
Zc3hav1
|
zinc finger CCCH type, antiviral 1 |
| chr5_-_121974873 | 1.03 |
ENSMUST00000118580.6
|
Sh2b3
|
SH2B adaptor protein 3 |
| chr2_+_143874979 | 1.01 |
ENSMUST00000037722.9
ENSMUST00000110032.2 |
Banf2
|
BANF family member 2 |
| chr4_-_56802266 | 0.99 |
ENSMUST00000030140.3
|
Elp1
|
elongator complex protein 1 |
| chr9_-_21913896 | 0.97 |
ENSMUST00000044926.6
|
Odad3
|
outer dynein arm docking complex subunit 3 |
| chrX_-_59449137 | 0.97 |
ENSMUST00000033480.13
ENSMUST00000101527.3 |
Atp11c
|
ATPase, class VI, type 11C |
| chr8_-_78244412 | 0.94 |
ENSMUST00000210922.2
ENSMUST00000210519.2 |
Arhgap10
|
Rho GTPase activating protein 10 |
| chr14_+_33675934 | 0.93 |
ENSMUST00000035695.10
|
Rbp3
|
retinol binding protein 3, interstitial |
| chr11_-_3881960 | 0.93 |
ENSMUST00000109990.8
|
Tcn2
|
transcobalamin 2 |
| chr1_-_74076279 | 0.93 |
ENSMUST00000187281.7
|
Tns1
|
tensin 1 |
| chr6_-_25809188 | 0.91 |
ENSMUST00000115327.8
|
Pot1a
|
protection of telomeres 1A |
| chr2_+_29951859 | 0.91 |
ENSMUST00000102866.10
|
Set
|
SET nuclear oncogene |
| chr7_+_16807965 | 0.88 |
ENSMUST00000071399.13
ENSMUST00000118367.3 |
Psg16
|
pregnancy specific glycoprotein 16 |
| chr1_-_44140820 | 0.87 |
ENSMUST00000152239.8
|
Tex30
|
testis expressed 30 |
| chr5_-_134935579 | 0.87 |
ENSMUST00000201316.4
ENSMUST00000047305.6 |
Tmem270
|
transmembrane protein 270 |
| chr2_+_126922156 | 0.87 |
ENSMUST00000142737.3
|
Blvra
|
biliverdin reductase A |
| chr10_-_40759307 | 0.83 |
ENSMUST00000044166.9
|
Cdc40
|
cell division cycle 40 |
| chr7_+_139900771 | 0.81 |
ENSMUST00000214594.2
|
Olfr525
|
olfactory receptor 525 |
| chrX_+_142936691 | 0.79 |
ENSMUST00000135687.2
|
A730046J19Rik
|
RIKEN cDNA A730046J19 gene |
| chr15_-_103473481 | 0.77 |
ENSMUST00000228060.2
ENSMUST00000228895.2 ENSMUST00000023134.5 |
Glycam1
|
glycosylation dependent cell adhesion molecule 1 |
| chr9_-_21913833 | 0.76 |
ENSMUST00000115336.10
|
Odad3
|
outer dynein arm docking complex subunit 3 |
| chr2_-_69416365 | 0.76 |
ENSMUST00000100051.9
ENSMUST00000092551.5 ENSMUST00000080953.12 |
Lrp2
|
low density lipoprotein receptor-related protein 2 |
| chr4_+_132262853 | 0.74 |
ENSMUST00000094657.10
ENSMUST00000105940.10 ENSMUST00000105939.10 ENSMUST00000150207.8 |
Dnajc8
|
DnaJ heat shock protein family (Hsp40) member C8 |
| chr4_+_155874896 | 0.73 |
ENSMUST00000165000.8
|
Ankrd65
|
ankyrin repeat domain 65 |
| chr10_+_13376745 | 0.73 |
ENSMUST00000060212.13
ENSMUST00000121465.3 |
Fuca2
|
fucosidase, alpha-L- 2, plasma |
| chr8_+_19545113 | 0.72 |
ENSMUST00000047851.4
|
Defb7
|
defensin beta 7 |
| chr5_+_115983292 | 0.72 |
ENSMUST00000137952.8
ENSMUST00000148245.8 |
Cit
|
citron |
| chr3_+_116653113 | 0.69 |
ENSMUST00000040260.11
|
Frrs1
|
ferric-chelate reductase 1 |
| chr17_-_35265514 | 0.69 |
ENSMUST00000007250.14
|
Msh5
|
mutS homolog 5 |
| chr6_-_57957795 | 0.68 |
ENSMUST00000176572.3
|
Vmn1r25
|
vomeronasal 1 receptor 25 |
| chr13_+_55740948 | 0.66 |
ENSMUST00000109905.5
|
Tmed9
|
transmembrane p24 trafficking protein 9 |
| chr2_-_152857239 | 0.65 |
ENSMUST00000028972.9
|
Pdrg1
|
p53 and DNA damage regulated 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.5 | 34.0 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 6.1 | 30.4 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 3.6 | 54.4 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 1.9 | 7.7 | GO:0071727 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 1.8 | 5.5 | GO:0072248 | metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 1.8 | 7.3 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 1.8 | 10.6 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 1.7 | 33.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 1.5 | 17.5 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 1.4 | 16.4 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 1.3 | 4.0 | GO:0034118 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 1.2 | 21.8 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 1.2 | 7.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 1.2 | 3.5 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 1.1 | 6.9 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.9 | 2.8 | GO:0046223 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) |
| 0.9 | 20.4 | GO:0038065 | collagen-activated signaling pathway(GO:0038065) |
| 0.9 | 2.6 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.9 | 2.6 | GO:1900135 | positive regulation of eosinophil degranulation(GO:0043311) regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) positive regulation of renin secretion into blood stream(GO:1900135) positive regulation of eosinophil activation(GO:1902568) |
| 0.8 | 3.4 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.8 | 4.2 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.8 | 21.8 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.8 | 19.3 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.7 | 4.7 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.7 | 2.0 | GO:0051977 | lysophospholipid transport(GO:0051977) |
| 0.6 | 3.9 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) positive regulation of somatostatin secretion(GO:0090274) |
| 0.6 | 8.4 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.6 | 10.7 | GO:0015816 | glycine transport(GO:0015816) |
| 0.5 | 2.1 | GO:0051316 | attachment of spindle microtubules to kinetochore involved in meiotic chromosome segregation(GO:0051316) |
| 0.5 | 4.9 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.5 | 19.1 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.5 | 16.6 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.4 | 12.6 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) |
| 0.4 | 4.3 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.4 | 1.3 | GO:0006059 | hexitol metabolic process(GO:0006059) naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 0.4 | 3.2 | GO:0008228 | opsonization(GO:0008228) |
| 0.4 | 4.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.4 | 3.6 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.3 | 3.3 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.3 | 1.6 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.3 | 1.8 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.3 | 8.6 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.3 | 4.6 | GO:0019374 | cellular defense response(GO:0006968) galactolipid metabolic process(GO:0019374) |
| 0.3 | 2.4 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.3 | 14.4 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.2 | 3.9 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.2 | 2.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.2 | 7.9 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.2 | 3.7 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.2 | 9.0 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.2 | 9.1 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.2 | 9.7 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.2 | 3.6 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.2 | 0.6 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.2 | 4.1 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.2 | 11.7 | GO:0070232 | regulation of T cell apoptotic process(GO:0070232) |
| 0.2 | 11.0 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) |
| 0.2 | 2.5 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.2 | 8.1 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.2 | 6.6 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.2 | 0.7 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.2 | 3.0 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.2 | 0.9 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 2.2 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 12.8 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 1.6 | GO:0007320 | insemination(GO:0007320) |
| 0.1 | 4.1 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.1 | 5.0 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 5.3 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.1 | 17.4 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 17.2 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.6 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.1 | 0.9 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 1.9 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.1 | 1.0 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 1.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 2.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 1.4 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.1 | 0.5 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 6.0 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.1 | 1.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 | 1.0 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.1 | 1.0 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.1 | 2.0 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.1 | 1.6 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 0.7 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 6.4 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 0.4 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 0.2 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.1 | 1.7 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 13.6 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 0.2 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.1 | 0.5 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 2.1 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.8 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 2.0 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.0 | 3.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 2.8 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 2.0 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.3 | GO:0060460 | left lung morphogenesis(GO:0060460) |
| 0.0 | 4.9 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 0.6 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.3 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 1.1 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.7 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 2.2 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 1.4 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 1.1 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 36.8 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 2.2 | 15.7 | GO:0032437 | cuticular plate(GO:0032437) |
| 1.6 | 4.8 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 1.6 | 4.7 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 1.0 | 12.6 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.7 | 2.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.6 | 10.6 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.5 | 7.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.4 | 8.5 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.4 | 21.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.4 | 2.9 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.4 | 1.8 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.3 | 1.4 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.3 | 3.4 | GO:0070187 | telosome(GO:0070187) |
| 0.3 | 57.7 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.3 | 4.6 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.3 | 1.8 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.3 | 1.3 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.2 | 5.0 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 0.9 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.2 | 4.6 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 6.8 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 17.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 1.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 3.4 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 53.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.6 | GO:0000802 | transverse filament(GO:0000802) |
| 0.1 | 18.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 2.9 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 1.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.7 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 12.8 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 1.7 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.1 | 104.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 4.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.1 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 4.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 2.1 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 3.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 2.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 57.1 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 3.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 28.8 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.7 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 2.6 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 15.0 | GO:0031410 | cytoplasmic vesicle(GO:0031410) |
| 0.0 | 2.2 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.7 | GO:0045171 | intercellular bridge(GO:0045171) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 30.4 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 3.6 | 21.8 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 2.0 | 4.0 | GO:0048030 | disaccharide binding(GO:0048030) |
| 1.8 | 10.6 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 1.6 | 4.8 | GO:0042497 | triacyl lipopeptide binding(GO:0042497) |
| 1.4 | 14.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 1.4 | 6.8 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 1.3 | 26.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 1.1 | 3.4 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 1.0 | 35.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 1.0 | 32.4 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.9 | 54.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.9 | 12.6 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.9 | 4.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.8 | 2.4 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.8 | 7.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.8 | 8.5 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.8 | 10.7 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.8 | 9.1 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.7 | 9.0 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.7 | 7.3 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.7 | 2.9 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.7 | 2.6 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.6 | 1.2 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.5 | 5.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.5 | 8.8 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.5 | 62.2 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.4 | 3.1 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.4 | 3.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.4 | 5.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.4 | 4.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.3 | 3.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.3 | 2.2 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.3 | 3.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.3 | 4.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.3 | 1.9 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.3 | 1.8 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.3 | 0.8 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.2 | 0.7 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.2 | 2.4 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 1.9 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.2 | 1.6 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.2 | 7.9 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.2 | 0.9 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.2 | 2.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 3.7 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 1.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.4 | GO:0051747 | cytosine C-5 DNA demethylase activity(GO:0051747) |
| 0.1 | 1.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 4.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.3 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 11.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 17.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 3.9 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 1.0 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 0.8 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.1 | 0.7 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.9 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 4.7 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.6 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 2.7 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.1 | 2.0 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 10.2 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 2.6 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 0.3 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.1 | 15.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 10.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 6.3 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 2.5 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.1 | 0.9 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.2 | GO:0015254 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.0 | 0.3 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 3.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 3.9 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.0 | 7.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 1.1 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 2.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 19.9 | GO:0000976 | transcription regulatory region sequence-specific DNA binding(GO:0000976) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 1.7 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 13.1 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 1.1 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 6.3 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.0 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 7.0 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 1.3 | GO:0000149 | SNARE binding(GO:0000149) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 11.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.5 | 25.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.5 | 7.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.5 | 32.8 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.3 | 10.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.3 | 1.9 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 6.0 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 7.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 9.1 | PID EPO PATHWAY | EPO signaling pathway |
| 0.2 | 4.7 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 4.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 24.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.6 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 4.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 9.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 3.2 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 2.2 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 3.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 4.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 2.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 4.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 30.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.6 | 21.8 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.6 | 7.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.6 | 9.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.5 | 4.8 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.5 | 20.7 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.5 | 37.0 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.4 | 21.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.4 | 6.8 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.3 | 5.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.3 | 18.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 49.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.3 | 8.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 3.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.2 | 12.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.2 | 7.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.2 | 5.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 2.9 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.2 | 9.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 4.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 4.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 0.5 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 2.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 9.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 2.4 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 2.8 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 5.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 2.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 2.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.8 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.6 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.8 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.6 | REACTOME MEIOSIS | Genes involved in Meiosis |