Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Tbp
Z-value: 3.70
Transcription factors associated with Tbp
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbp
|
ENSMUSG00000014767.18 | Tbp |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbp | mm39_v1_chr17_+_15720150_15720222 | -0.44 | 1.2e-04 | Click! |
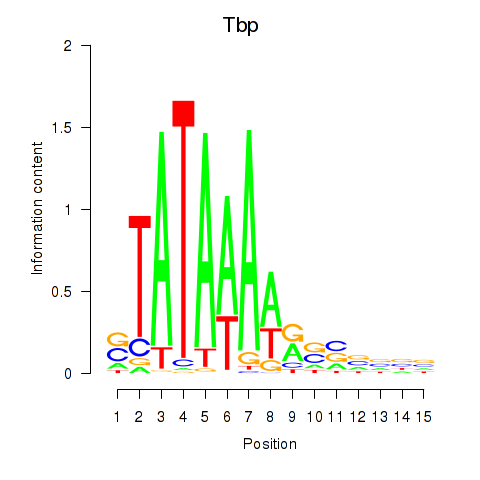
Activity profile of Tbp motif
Sorted Z-values of Tbp motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbp
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_94863825 | 45.00 |
ENSMUST00000034207.8
|
Mt4
|
metallothionein 4 |
| chr19_+_39049442 | 40.18 |
ENSMUST00000087236.5
|
Cyp2c65
|
cytochrome P450, family 2, subfamily c, polypeptide 65 |
| chr3_+_106020545 | 34.77 |
ENSMUST00000079132.12
ENSMUST00000139086.2 |
Chia1
|
chitinase, acidic 1 |
| chr11_-_100012384 | 33.18 |
ENSMUST00000007275.3
|
Krt13
|
keratin 13 |
| chr3_-_92366022 | 30.26 |
ENSMUST00000058142.4
|
Sprr3
|
small proline-rich protein 3 |
| chr6_-_113696390 | 28.03 |
ENSMUST00000203588.2
ENSMUST00000204163.3 ENSMUST00000203363.3 |
Ghrl
|
ghrelin |
| chr2_-_164197987 | 26.14 |
ENSMUST00000165980.2
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr6_-_113696809 | 24.25 |
ENSMUST00000203770.3
ENSMUST00000064993.8 |
Ghrl
|
ghrelin |
| chr17_-_31383976 | 23.78 |
ENSMUST00000235870.2
|
Tff1
|
trefoil factor 1 |
| chr9_+_98372575 | 23.65 |
ENSMUST00000035029.3
|
Rbp2
|
retinol binding protein 2, cellular |
| chr3_+_92851790 | 22.71 |
ENSMUST00000055375.6
|
Lce3c
|
late cornified envelope 3C |
| chr2_-_164198427 | 22.66 |
ENSMUST00000109367.10
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr15_-_101588714 | 22.62 |
ENSMUST00000023786.7
|
Krt6b
|
keratin 6B |
| chr15_-_101912917 | 22.29 |
ENSMUST00000023952.10
|
Krt8
|
keratin 8 |
| chr9_+_46151994 | 21.90 |
ENSMUST00000034585.7
|
Apoa4
|
apolipoprotein A-IV |
| chr16_+_48662894 | 21.67 |
ENSMUST00000238847.2
ENSMUST00000023329.7 |
Retnla
|
resistin like alpha |
| chr3_+_93301003 | 21.41 |
ENSMUST00000045912.3
|
Rptn
|
repetin |
| chr15_-_101621332 | 21.41 |
ENSMUST00000023709.7
|
Krt5
|
keratin 5 |
| chr3_-_92926364 | 20.77 |
ENSMUST00000193944.2
ENSMUST00000029520.9 |
Lce1m
|
late cornified envelope 1M |
| chr3_-_92734546 | 20.25 |
ENSMUST00000072363.5
|
Kprp
|
keratinocyte expressed, proline-rich |
| chr17_-_31348576 | 20.18 |
ENSMUST00000024827.5
|
Tff3
|
trefoil factor 3, intestinal |
| chr11_-_99328969 | 19.99 |
ENSMUST00000017743.3
|
Krt20
|
keratin 20 |
| chr8_+_118428643 | 19.93 |
ENSMUST00000034304.9
|
Hsd17b2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr11_-_100026754 | 19.32 |
ENSMUST00000107411.3
|
Krt15
|
keratin 15 |
| chr4_-_41098174 | 18.77 |
ENSMUST00000055327.8
|
Aqp3
|
aquaporin 3 |
| chr3_+_92899521 | 18.07 |
ENSMUST00000090863.5
|
Lce3f
|
late cornified envelope 3F |
| chr2_+_122479770 | 17.84 |
ENSMUST00000047498.15
ENSMUST00000110512.4 |
AA467197
|
expressed sequence AA467197 |
| chr11_+_94827050 | 17.63 |
ENSMUST00000001547.8
|
Col1a1
|
collagen, type I, alpha 1 |
| chr15_-_101759212 | 17.55 |
ENSMUST00000023790.5
|
Krt1
|
keratin 1 |
| chr7_-_30623592 | 17.20 |
ENSMUST00000217812.2
ENSMUST00000074671.9 |
Hamp2
|
hepcidin antimicrobial peptide 2 |
| chr16_-_48232770 | 17.03 |
ENSMUST00000212197.2
|
Gm5485
|
predicted gene 5485 |
| chr2_-_62313981 | 16.78 |
ENSMUST00000136686.2
ENSMUST00000102733.10 |
Gcg
|
glucagon |
| chr19_+_39102342 | 16.69 |
ENSMUST00000087234.3
|
Cyp2c66
|
cytochrome P450, family 2, subfamily c, polypeptide 66 |
| chr5_+_135916847 | 16.56 |
ENSMUST00000111155.2
|
Hspb1
|
heat shock protein 1 |
| chr3_-_92393193 | 16.42 |
ENSMUST00000054599.8
|
Sprr1a
|
small proline-rich protein 1A |
| chr5_+_135916764 | 16.36 |
ENSMUST00000005077.7
|
Hspb1
|
heat shock protein 1 |
| chr3_-_92777521 | 15.63 |
ENSMUST00000163439.3
|
2310050C09Rik
|
RIKEN cDNA 2310050C09 gene |
| chr3_+_137923521 | 15.61 |
ENSMUST00000090171.7
|
Adh7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr6_-_67512768 | 15.21 |
ENSMUST00000058178.6
|
Tacstd2
|
tumor-associated calcium signal transducer 2 |
| chr15_-_101602734 | 15.19 |
ENSMUST00000023788.8
|
Krt6a
|
keratin 6A |
| chr14_+_56279498 | 15.10 |
ENSMUST00000015576.6
|
Mcpt2
|
mast cell protease 2 |
| chr11_-_69838971 | 15.07 |
ENSMUST00000179298.3
ENSMUST00000018710.13 ENSMUST00000135437.3 ENSMUST00000141837.9 ENSMUST00000142500.8 |
Slc2a4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr2_+_122129379 | 14.68 |
ENSMUST00000028656.2
|
Duoxa2
|
dual oxidase maturation factor 2 |
| chr1_+_119934624 | 14.65 |
ENSMUST00000072886.11
ENSMUST00000189037.2 |
Sctr
|
secretin receptor |
| chr6_+_41331039 | 14.52 |
ENSMUST00000072103.7
|
Try10
|
trypsin 10 |
| chr5_+_115061293 | 14.48 |
ENSMUST00000031540.11
ENSMUST00000112143.4 |
Oasl1
|
2'-5' oligoadenylate synthetase-like 1 |
| chr3_-_92833537 | 14.15 |
ENSMUST00000067318.6
|
Lce3a
|
late cornified envelope 3A |
| chr3_-_92577621 | 13.73 |
ENSMUST00000029530.6
|
Lce1a2
|
late cornified envelope 1A2 |
| chr8_+_19248718 | 13.66 |
ENSMUST00000081017.3
|
Defb4
|
defensin beta 4 |
| chr3_-_90603013 | 13.57 |
ENSMUST00000069960.12
ENSMUST00000117167.2 |
S100a9
|
S100 calcium binding protein A9 (calgranulin B) |
| chr11_-_100151847 | 13.54 |
ENSMUST00000080893.7
|
Krt17
|
keratin 17 |
| chr7_-_18757461 | 13.15 |
ENSMUST00000036018.6
|
Foxa3
|
forkhead box A3 |
| chr7_+_130633776 | 12.70 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr3_+_92246440 | 12.62 |
ENSMUST00000047477.4
|
Sprr2d
|
small proline-rich protein 2D |
| chr7_+_30252687 | 12.49 |
ENSMUST00000044048.8
|
Hspb6
|
heat shock protein, alpha-crystallin-related, B6 |
| chr14_+_56255422 | 12.30 |
ENSMUST00000022836.6
|
Mcpt1
|
mast cell protease 1 |
| chr14_-_56299764 | 12.29 |
ENSMUST00000043249.10
|
Mcpt4
|
mast cell protease 4 |
| chr7_+_141276575 | 12.24 |
ENSMUST00000185406.8
|
Muc2
|
mucin 2 |
| chr3_+_92864693 | 12.22 |
ENSMUST00000059053.11
|
Lce3d
|
late cornified envelope 3D |
| chr13_-_73848807 | 12.17 |
ENSMUST00000022048.6
|
Slc6a19
|
solute carrier family 6 (neurotransmitter transporter), member 19 |
| chr11_-_98915005 | 12.13 |
ENSMUST00000068031.8
|
Top2a
|
topoisomerase (DNA) II alpha |
| chr3_-_20296337 | 11.97 |
ENSMUST00000001921.3
|
Cpa3
|
carboxypeptidase A3, mast cell |
| chr7_-_141241632 | 11.50 |
ENSMUST00000239500.1
|
ENSMUSG00000118661.1
|
mucin 6, gastric |
| chr11_-_101998648 | 11.22 |
ENSMUST00000177304.8
ENSMUST00000017455.15 |
Pyy
|
peptide YY |
| chr19_+_38995463 | 11.08 |
ENSMUST00000025966.5
|
Cyp2c55
|
cytochrome P450, family 2, subfamily c, polypeptide 55 |
| chr5_-_91550853 | 11.05 |
ENSMUST00000121044.6
|
Btc
|
betacellulin, epidermal growth factor family member |
| chr3_+_57332735 | 11.04 |
ENSMUST00000029377.8
|
Tm4sf4
|
transmembrane 4 superfamily member 4 |
| chr6_-_41291634 | 11.02 |
ENSMUST00000064324.12
|
Try5
|
trypsin 5 |
| chr9_-_67739607 | 11.02 |
ENSMUST00000054500.7
|
C2cd4a
|
C2 calcium-dependent domain containing 4A |
| chr10_+_96452860 | 10.97 |
ENSMUST00000038377.9
|
Btg1
|
BTG anti-proliferation factor 1 |
| chr9_-_95697441 | 10.84 |
ENSMUST00000119760.2
|
Pls1
|
plastin 1 (I-isoform) |
| chr2_+_152578164 | 10.84 |
ENSMUST00000038368.9
ENSMUST00000109824.2 |
Id1
|
inhibitor of DNA binding 1, HLH protein |
| chr1_-_37996838 | 10.67 |
ENSMUST00000027254.10
ENSMUST00000114894.2 |
Lyg1
|
lysozyme G-like 1 |
| chr10_-_80512117 | 10.65 |
ENSMUST00000200082.5
|
Mknk2
|
MAP kinase-interacting serine/threonine kinase 2 |
| chr15_+_10952418 | 10.44 |
ENSMUST00000022853.15
ENSMUST00000110523.2 |
C1qtnf3
|
C1q and tumor necrosis factor related protein 3 |
| chr5_+_90920294 | 10.00 |
ENSMUST00000031320.8
|
Pf4
|
platelet factor 4 |
| chr4_-_57956411 | 9.99 |
ENSMUST00000030051.6
|
Txn1
|
thioredoxin 1 |
| chr11_+_94455865 | 9.98 |
ENSMUST00000040418.9
|
Chad
|
chondroadherin |
| chr14_-_52011035 | 9.96 |
ENSMUST00000073860.6
|
Ang4
|
angiogenin, ribonuclease A family, member 4 |
| chr5_+_90920353 | 9.90 |
ENSMUST00000202625.2
|
Pf4
|
platelet factor 4 |
| chr14_+_69846517 | 9.85 |
ENSMUST00000022660.14
|
Loxl2
|
lysyl oxidase-like 2 |
| chr7_-_126651847 | 9.76 |
ENSMUST00000205424.2
|
Zg16
|
zymogen granule protein 16 |
| chr2_+_129854256 | 9.46 |
ENSMUST00000110299.3
|
Tgm3
|
transglutaminase 3, E polypeptide |
| chr11_+_33913013 | 9.25 |
ENSMUST00000020362.3
|
Kcnmb1
|
potassium large conductance calcium-activated channel, subfamily M, beta member 1 |
| chr1_-_134006847 | 9.05 |
ENSMUST00000020692.7
|
Btg2
|
BTG anti-proliferation factor 2 |
| chr11_-_120441952 | 8.97 |
ENSMUST00000026121.3
|
Ppp1r27
|
protein phosphatase 1, regulatory subunit 27 |
| chr14_-_47426863 | 8.88 |
ENSMUST00000089959.7
|
Gch1
|
GTP cyclohydrolase 1 |
| chr2_+_164174660 | 8.73 |
ENSMUST00000017148.8
|
Svs5
|
seminal vesicle secretory protein 5 |
| chr4_+_120523758 | 8.71 |
ENSMUST00000094814.6
|
Cited4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chr7_-_44181477 | 8.58 |
ENSMUST00000098483.9
ENSMUST00000035323.6 |
Spib
|
Spi-B transcription factor (Spi-1/PU.1 related) |
| chr7_-_15993110 | 8.50 |
ENSMUST00000168818.2
|
C5ar1
|
complement component 5a receptor 1 |
| chr15_-_101801351 | 8.48 |
ENSMUST00000100179.2
|
Krt76
|
keratin 76 |
| chr12_+_8027640 | 8.46 |
ENSMUST00000171271.8
ENSMUST00000037811.13 |
Apob
|
apolipoprotein B |
| chr1_-_36312482 | 8.21 |
ENSMUST00000056946.8
|
Neurl3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr2_-_164120215 | 8.02 |
ENSMUST00000017142.3
|
Svs4
|
seminal vesicle secretory protein 4 |
| chr12_+_8027767 | 7.92 |
ENSMUST00000037520.14
|
Apob
|
apolipoprotein B |
| chr15_-_101482320 | 7.72 |
ENSMUST00000042957.6
|
Krt75
|
keratin 75 |
| chr17_-_34174631 | 7.64 |
ENSMUST00000174609.9
ENSMUST00000008812.9 |
Rps18
|
ribosomal protein S18 |
| chr9_+_7272514 | 7.46 |
ENSMUST00000015394.10
|
Mmp13
|
matrix metallopeptidase 13 |
| chrX_+_73314418 | 7.40 |
ENSMUST00000008826.14
ENSMUST00000151702.8 ENSMUST00000074085.12 ENSMUST00000135690.2 |
Rpl10
|
ribosomal protein L10 |
| chr17_-_84495364 | 7.35 |
ENSMUST00000060366.7
|
Zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr17_-_57535003 | 7.24 |
ENSMUST00000177046.2
ENSMUST00000024988.15 |
C3
|
complement component 3 |
| chr15_-_101555815 | 7.20 |
ENSMUST00000229963.2
ENSMUST00000100184.4 |
Gm5478
|
predicted pseudogene 5478 |
| chr17_+_24939072 | 6.95 |
ENSMUST00000054289.13
|
Rps2
|
ribosomal protein S2 |
| chr11_-_53313950 | 6.84 |
ENSMUST00000036045.6
|
Leap2
|
liver-expressed antimicrobial peptide 2 |
| chr11_-_99519057 | 6.82 |
ENSMUST00000081007.7
|
Krtap4-1
|
keratin associated protein 4-1 |
| chr2_+_13579092 | 6.80 |
ENSMUST00000193675.2
|
Vim
|
vimentin |
| chr3_+_87855973 | 6.76 |
ENSMUST00000005019.6
|
Crabp2
|
cellular retinoic acid binding protein II |
| chr11_+_76889415 | 6.67 |
ENSMUST00000108402.9
ENSMUST00000021195.11 |
Slc6a4
|
solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 |
| chr6_+_72575458 | 6.57 |
ENSMUST00000070597.13
ENSMUST00000176364.8 ENSMUST00000176168.3 |
Retsat
|
retinol saturase (all trans retinol 13,14 reductase) |
| chr3_+_129630380 | 6.53 |
ENSMUST00000077918.7
|
Cfi
|
complement component factor i |
| chr17_+_24939037 | 6.43 |
ENSMUST00000170715.8
|
Rps2
|
ribosomal protein S2 |
| chr1_-_30988772 | 6.40 |
ENSMUST00000238874.2
ENSMUST00000027232.15 ENSMUST00000076587.6 ENSMUST00000233506.2 |
Ptp4a1
|
protein tyrosine phosphatase 4a1 |
| chr6_+_78402956 | 6.37 |
ENSMUST00000079926.6
|
Reg1
|
regenerating islet-derived 1 |
| chr2_+_164158651 | 6.35 |
ENSMUST00000017144.3
|
Svs6
|
seminal vesicle secretory protein 6 |
| chr18_+_34757666 | 6.33 |
ENSMUST00000167161.9
|
Kif20a
|
kinesin family member 20A |
| chrX_+_140258381 | 6.31 |
ENSMUST00000112931.8
ENSMUST00000112930.8 |
Col4a5
|
collagen, type IV, alpha 5 |
| chr12_+_102521225 | 6.27 |
ENSMUST00000021610.7
|
Chga
|
chromogranin A |
| chr18_+_44237577 | 6.26 |
ENSMUST00000239465.2
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr1_-_37580084 | 6.10 |
ENSMUST00000151952.8
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr9_+_7502341 | 6.03 |
ENSMUST00000034488.4
|
Mmp10
|
matrix metallopeptidase 10 |
| chr18_+_34758062 | 6.03 |
ENSMUST00000166044.3
|
Kif20a
|
kinesin family member 20A |
| chr11_-_115405200 | 6.01 |
ENSMUST00000021083.7
|
Jpt1
|
Jupiter microtubule associated homolog 1 |
| chr7_-_140846328 | 6.00 |
ENSMUST00000106023.8
ENSMUST00000097952.9 ENSMUST00000026571.11 |
Irf7
|
interferon regulatory factor 7 |
| chr16_-_10609959 | 5.99 |
ENSMUST00000037996.7
|
Prm2
|
protamine 2 |
| chr7_-_143056252 | 5.99 |
ENSMUST00000010904.5
|
Phlda2
|
pleckstrin homology like domain, family A, member 2 |
| chr3_-_20329823 | 5.96 |
ENSMUST00000011607.6
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr16_-_55642491 | 5.90 |
ENSMUST00000114458.8
|
Nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta |
| chr17_-_14914484 | 5.88 |
ENSMUST00000170872.3
|
Thbs2
|
thrombospondin 2 |
| chr3_+_92259448 | 5.82 |
ENSMUST00000068399.2
|
Sprr2e
|
small proline-rich protein 2E |
| chr6_-_129077867 | 5.80 |
ENSMUST00000032258.8
|
Clec2e
|
C-type lectin domain family 2, member e |
| chr8_-_13304154 | 5.69 |
ENSMUST00000204916.3
ENSMUST00000033825.11 |
Adprhl1
|
ADP-ribosylhydrolase like 1 |
| chr11_+_67167950 | 5.61 |
ENSMUST00000019625.12
|
Myh8
|
myosin, heavy polypeptide 8, skeletal muscle, perinatal |
| chr8_+_85696396 | 5.58 |
ENSMUST00000109733.8
|
Prdx2
|
peroxiredoxin 2 |
| chr17_-_16051295 | 5.56 |
ENSMUST00000231985.2
|
Rgmb
|
repulsive guidance molecule family member B |
| chr17_+_24939225 | 5.48 |
ENSMUST00000146867.2
|
Rps2
|
ribosomal protein S2 |
| chr3_+_145855929 | 5.47 |
ENSMUST00000098524.5
|
Mcoln2
|
mucolipin 2 |
| chr1_-_66974492 | 5.45 |
ENSMUST00000120415.8
ENSMUST00000119429.8 |
Myl1
|
myosin, light polypeptide 1 |
| chr18_+_44237474 | 5.38 |
ENSMUST00000081271.7
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr5_+_24633206 | 5.33 |
ENSMUST00000115049.9
|
Slc4a2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr4_+_135870808 | 5.27 |
ENSMUST00000008016.3
|
Id3
|
inhibitor of DNA binding 3 |
| chr6_-_136852792 | 5.08 |
ENSMUST00000032342.3
|
Mgp
|
matrix Gla protein |
| chr17_-_34043502 | 5.02 |
ENSMUST00000087342.13
ENSMUST00000173844.8 |
Rps28
|
ribosomal protein S28 |
| chr3_-_92346078 | 5.02 |
ENSMUST00000062160.4
|
Sprr1b
|
small proline-rich protein 1B |
| chr7_-_46392403 | 4.98 |
ENSMUST00000128088.4
|
Saa1
|
serum amyloid A 1 |
| chr11_+_99755302 | 4.98 |
ENSMUST00000092694.4
|
Gm11559
|
predicted gene 11559 |
| chr14_+_29744590 | 4.85 |
ENSMUST00000118917.2
|
Chdh
|
choline dehydrogenase |
| chr17_-_35293447 | 4.84 |
ENSMUST00000007259.4
|
Ly6g6d
|
lymphocyte antigen 6 complex, locus G6D |
| chr15_-_5093222 | 4.84 |
ENSMUST00000110689.5
|
C7
|
complement component 7 |
| chr17_-_34043320 | 4.83 |
ENSMUST00000173879.8
ENSMUST00000166693.3 ENSMUST00000173019.8 |
Rps28
|
ribosomal protein S28 |
| chr7_+_119773070 | 4.78 |
ENSMUST00000033201.7
|
Anks4b
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr10_-_128462616 | 4.72 |
ENSMUST00000026420.7
|
Rps26
|
ribosomal protein S26 |
| chr2_-_164080358 | 4.72 |
ENSMUST00000044953.3
|
Svs2
|
seminal vesicle secretory protein 2 |
| chr16_-_55659194 | 4.70 |
ENSMUST00000096026.9
ENSMUST00000036273.13 ENSMUST00000114457.8 |
Nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta |
| chr8_+_85696453 | 4.65 |
ENSMUST00000125893.8
|
Prdx2
|
peroxiredoxin 2 |
| chr13_+_23947641 | 4.63 |
ENSMUST00000055770.4
|
H1f1
|
H1.1 linker histone, cluster member |
| chr6_+_29768470 | 4.52 |
ENSMUST00000102995.9
ENSMUST00000115242.9 |
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr14_-_56339915 | 4.52 |
ENSMUST00000015583.2
|
Ctsg
|
cathepsin G |
| chr11_-_83469446 | 4.40 |
ENSMUST00000019266.6
|
Ccl9
|
chemokine (C-C motif) ligand 9 |
| chr6_-_88604404 | 4.38 |
ENSMUST00000120933.5
|
Kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr17_+_80514889 | 4.36 |
ENSMUST00000134652.2
|
Ttc39d
|
tetratricopeptide repeat domain 39D |
| chr5_-_135601887 | 4.36 |
ENSMUST00000004936.10
ENSMUST00000201401.2 |
Ccl24
|
chemokine (C-C motif) ligand 24 |
| chr6_-_8180174 | 4.33 |
ENSMUST00000213284.2
|
Col28a1
|
collagen, type XXVIII, alpha 1 |
| chr13_-_3943556 | 4.31 |
ENSMUST00000099946.6
|
Net1
|
neuroepithelial cell transforming gene 1 |
| chr11_-_99482165 | 4.28 |
ENSMUST00000104930.2
|
Krtap1-3
|
keratin associated protein 1-3 |
| chr2_-_93787441 | 4.27 |
ENSMUST00000099689.5
|
Gm13889
|
predicted gene 13889 |
| chr8_+_62381115 | 4.24 |
ENSMUST00000154398.8
ENSMUST00000156980.8 ENSMUST00000093485.3 ENSMUST00000070631.15 |
Ddx60
|
DExD/H box helicase 60 |
| chr2_+_36120438 | 4.12 |
ENSMUST00000062069.6
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr13_+_23868175 | 3.98 |
ENSMUST00000018246.6
|
H2bc4
|
H2B clustered histone 4 |
| chr15_-_65784103 | 3.98 |
ENSMUST00000079776.14
|
Oc90
|
otoconin 90 |
| chr11_+_87684299 | 3.97 |
ENSMUST00000020779.11
|
Mpo
|
myeloperoxidase |
| chr11_-_57722830 | 3.97 |
ENSMUST00000036917.3
|
Hand1
|
heart and neural crest derivatives expressed 1 |
| chr16_-_10614679 | 3.95 |
ENSMUST00000023144.6
|
Prm1
|
protamine 1 |
| chr8_-_3798941 | 3.90 |
ENSMUST00000012847.3
|
Cd209a
|
CD209a antigen |
| chr8_-_13304068 | 3.90 |
ENSMUST00000168498.8
|
Adprhl1
|
ADP-ribosylhydrolase like 1 |
| chr10_+_21870565 | 3.88 |
ENSMUST00000020145.12
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr15_-_102154874 | 3.87 |
ENSMUST00000063339.14
|
Rarg
|
retinoic acid receptor, gamma |
| chr3_-_51248032 | 3.83 |
ENSMUST00000062009.14
ENSMUST00000194641.6 |
Elf2
|
E74-like factor 2 |
| chr11_-_33113071 | 3.81 |
ENSMUST00000093201.13
ENSMUST00000101375.5 ENSMUST00000109354.10 ENSMUST00000075641.10 |
Npm1
|
nucleophosmin 1 |
| chr6_+_68402550 | 3.80 |
ENSMUST00000103323.3
|
Igkv16-104
|
immunoglobulin kappa variable 16-104 |
| chr10_+_51367052 | 3.79 |
ENSMUST00000217705.2
ENSMUST00000078778.5 ENSMUST00000220182.2 ENSMUST00000220226.2 |
Lilrb4a
|
leukocyte immunoglobulin-like receptor, subfamily B, member 4A |
| chr6_-_50631418 | 3.75 |
ENSMUST00000031853.8
|
Npvf
|
neuropeptide VF precursor |
| chr2_-_164080172 | 3.74 |
ENSMUST00000109374.2
|
Svs2
|
seminal vesicle secretory protein 2 |
| chr4_-_3835595 | 3.72 |
ENSMUST00000138502.2
|
Rps20
|
ribosomal protein S20 |
| chr2_+_119449192 | 3.69 |
ENSMUST00000028771.8
|
Nusap1
|
nucleolar and spindle associated protein 1 |
| chr7_-_102408573 | 3.68 |
ENSMUST00000210453.3
ENSMUST00000232246.3 ENSMUST00000239110.2 ENSMUST00000060187.15 ENSMUST00000168007.3 |
Olfr560
Olfr78
|
olfactory receptor 560 olfactory receptor 78 |
| chr3_-_105940130 | 3.68 |
ENSMUST00000200146.2
|
Chil5
|
chitinase-like 5 |
| chr1_-_155617773 | 3.68 |
ENSMUST00000027740.14
|
Lhx4
|
LIM homeobox protein 4 |
| chr11_-_77616103 | 3.65 |
ENSMUST00000078623.5
|
Cryba1
|
crystallin, beta A1 |
| chr19_-_3464447 | 3.64 |
ENSMUST00000025842.8
ENSMUST00000237521.2 |
Gal
|
galanin and GMAP prepropeptide |
| chr15_-_65784246 | 3.62 |
ENSMUST00000060522.11
|
Oc90
|
otoconin 90 |
| chr6_-_41354538 | 3.61 |
ENSMUST00000096003.7
|
Prss3
|
protease, serine 3 |
| chr8_-_13304096 | 3.60 |
ENSMUST00000171619.2
|
Adprhl1
|
ADP-ribosylhydrolase like 1 |
| chrX_+_163221035 | 3.58 |
ENSMUST00000033755.6
|
Asb11
|
ankyrin repeat and SOCS box-containing 11 |
| chr12_+_85520652 | 3.58 |
ENSMUST00000021674.7
|
Fos
|
FBJ osteosarcoma oncogene |
| chr16_+_65612394 | 3.58 |
ENSMUST00000227997.2
|
Vgll3
|
vestigial like family member 3 |
| chr8_+_22108199 | 3.56 |
ENSMUST00000074343.6
|
Defa26
|
defensin, alpha, 26 |
| chr2_+_158099986 | 3.55 |
ENSMUST00000109499.8
ENSMUST00000109500.8 ENSMUST00000065039.3 |
Bpi
|
bactericidal permeablility increasing protein |
| chr1_+_130947580 | 3.54 |
ENSMUST00000016673.6
|
Il10
|
interleukin 10 |
| chr9_+_55448432 | 3.52 |
ENSMUST00000034869.11
|
Isl2
|
insulin related protein 2 (islet 2) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.1 | 52.3 | GO:1904346 | regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) negative regulation of energy homeostasis(GO:2000506) |
| 7.3 | 21.9 | GO:0034443 | negative regulation of lipoprotein oxidation(GO:0034443) multicellular organism lipid catabolic process(GO:0044240) |
| 6.3 | 18.8 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 5.6 | 22.3 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 5.1 | 15.2 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 4.9 | 14.8 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 4.3 | 38.5 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 4.2 | 21.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 3.9 | 15.6 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 3.7 | 32.9 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 3.3 | 13.1 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 3.3 | 133.5 | GO:0031424 | keratinization(GO:0031424) |
| 3.1 | 9.3 | GO:1903413 | response to bile acid(GO:1903412) cellular response to bile acid(GO:1903413) |
| 2.7 | 10.8 | GO:1902896 | terminal web assembly(GO:1902896) |
| 2.6 | 23.7 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 2.4 | 12.1 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 2.4 | 134.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 2.3 | 27.6 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 2.2 | 8.9 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 2.2 | 19.9 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 2.1 | 8.5 | GO:0034760 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 2.1 | 16.8 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 1.9 | 3.8 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 1.9 | 13.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 1.9 | 24.3 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 1.9 | 7.5 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 1.8 | 14.7 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 1.8 | 7.2 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 1.8 | 63.3 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 1.7 | 6.7 | GO:0021941 | negative regulation of cerebellar granule cell precursor proliferation(GO:0021941) |
| 1.6 | 14.8 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 1.6 | 6.3 | GO:1901079 | positive regulation of relaxation of muscle(GO:1901079) regulation of dense core granule biogenesis(GO:2000705) |
| 1.5 | 4.4 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 1.5 | 4.4 | GO:0060720 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 1.4 | 1.4 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 1.4 | 11.0 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 1.3 | 4.0 | GO:0002149 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 1.3 | 10.4 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 1.3 | 15.5 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 1.3 | 10.1 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 1.3 | 10.0 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 1.3 | 41.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 1.2 | 3.7 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 1.2 | 8.5 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 1.2 | 3.6 | GO:0051794 | regulation of catagen(GO:0051794) positive regulation of catagen(GO:0051795) |
| 1.2 | 3.5 | GO:0060300 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) regulation of cytokine activity(GO:0060300) |
| 1.2 | 3.5 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 1.2 | 16.4 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 1.1 | 28.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 1.1 | 5.6 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 1.0 | 48.6 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 1.0 | 6.0 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 1.0 | 4.8 | GO:1904970 | brush border assembly(GO:1904970) |
| 1.0 | 2.9 | GO:0045297 | mating plug formation(GO:0042628) single-organism reproductive behavior(GO:0044704) post-mating behavior(GO:0045297) |
| 0.9 | 3.7 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.9 | 10.0 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.9 | 3.5 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.9 | 15.6 | GO:0032096 | negative regulation of response to food(GO:0032096) |
| 0.8 | 7.4 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.8 | 4.9 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.8 | 3.9 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.8 | 12.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.8 | 2.3 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.8 | 10.7 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.8 | 0.8 | GO:0090420 | naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 0.7 | 12.6 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.7 | 2.9 | GO:0002930 | trabecular meshwork development(GO:0002930) |
| 0.7 | 2.9 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.7 | 2.9 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.7 | 12.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.7 | 4.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.7 | 5.6 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.7 | 3.5 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.7 | 2.8 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.7 | 4.7 | GO:0051105 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.7 | 15.5 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.6 | 3.7 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.6 | 4.2 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.5 | 69.4 | GO:0007586 | digestion(GO:0007586) |
| 0.5 | 14.0 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.5 | 3.7 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.5 | 6.8 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.5 | 2.1 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.5 | 4.5 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.5 | 9.6 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.5 | 3.2 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.4 | 5.2 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.4 | 2.2 | GO:0035747 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.4 | 9.1 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.4 | 1.3 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.4 | 2.1 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.4 | 2.4 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.4 | 1.6 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.4 | 9.0 | GO:0033275 | actin-myosin filament sliding(GO:0033275) |
| 0.4 | 15.1 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.4 | 8.5 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.3 | 1.0 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.3 | 1.4 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.3 | 14.3 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.3 | 2.0 | GO:0032275 | luteinizing hormone secretion(GO:0032275) |
| 0.3 | 2.2 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.3 | 3.1 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.3 | 3.1 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.3 | 12.5 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.3 | 3.0 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.3 | 1.8 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.3 | 6.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.3 | 15.5 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.3 | 9.7 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.3 | 9.9 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.3 | 4.6 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.3 | 0.8 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.3 | 1.8 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.3 | 9.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 7.6 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.2 | 3.9 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.2 | 4.3 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.2 | 3.3 | GO:2000018 | regulation of male gonad development(GO:2000018) |
| 0.2 | 1.1 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.2 | 14.4 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.2 | 3.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.2 | 8.5 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.2 | 8.6 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.2 | 10.0 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.2 | 0.8 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.2 | 1.7 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.2 | 3.2 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.2 | 1.7 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.2 | 3.4 | GO:0030802 | regulation of cyclic nucleotide biosynthetic process(GO:0030802) |
| 0.2 | 1.2 | GO:0002370 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.2 | 1.0 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.2 | 4.8 | GO:1905145 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.2 | 13.2 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.2 | 1.8 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 7.3 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.1 | 7.1 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.1 | 2.0 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 1.1 | GO:0060297 | regulation of sarcomere organization(GO:0060297) |
| 0.1 | 2.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 1.8 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 2.9 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 12.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 0.7 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 1.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 6.4 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 7.0 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.1 | 2.3 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.1 | 0.3 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.1 | 5.1 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.1 | 0.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 1.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 7.1 | GO:0043627 | response to estrogen(GO:0043627) |
| 0.1 | 0.5 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 0.1 | GO:1901536 | negative regulation of DNA demethylation(GO:1901536) |
| 0.1 | 2.2 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.1 | 10.0 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.1 | 1.0 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 6.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 5.1 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.6 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 2.0 | GO:0007595 | lactation(GO:0007595) |
| 0.0 | 0.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 6.5 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 6.2 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 3.0 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.4 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.0 | 0.8 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 2.7 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 6.0 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.6 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 4.8 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 3.1 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 1.6 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 2.4 | GO:0006497 | protein lipidation(GO:0006497) |
| 0.0 | 4.4 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.2 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.0 | 1.4 | GO:0030193 | regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.5 | GO:0060713 | labyrinthine layer morphogenesis(GO:0060713) labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 4.4 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 2.4 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 2.8 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.0 | 1.0 | GO:0042107 | cytokine metabolic process(GO:0042107) |
| 0.0 | 0.5 | GO:0060043 | regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.0 | 0.6 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 4.0 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 17.6 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 5.5 | 16.4 | GO:0034359 | mature chylomicron(GO:0034359) |
| 4.3 | 238.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 2.3 | 99.0 | GO:0045095 | keratin filament(GO:0045095) |
| 1.4 | 27.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 1.4 | 19.9 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 1.2 | 10.8 | GO:1990357 | terminal web(GO:1990357) |
| 1.1 | 3.4 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 1.0 | 6.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 1.0 | 10.0 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 1.0 | 12.7 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.9 | 17.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.9 | 6.3 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.9 | 9.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.8 | 15.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.7 | 44.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.7 | 3.6 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.5 | 4.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.5 | 9.9 | GO:0042627 | chylomicron(GO:0042627) |
| 0.5 | 1.4 | GO:0060473 | cortical granule(GO:0060473) |
| 0.4 | 1.8 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.4 | 2.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.4 | 7.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.3 | 10.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.3 | 1.3 | GO:1990589 | Lewy body core(GO:1990037) ATF4-CREB1 transcription factor complex(GO:1990589) ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.3 | 1.8 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.3 | 6.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.3 | 3.1 | GO:0005767 | secondary lysosome(GO:0005767) autolysosome(GO:0044754) |
| 0.3 | 2.2 | GO:0001652 | granular component(GO:0001652) |
| 0.3 | 1.8 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.2 | 10.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 1.4 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.2 | 4.0 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.2 | 2.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.2 | 6.4 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.2 | 5.9 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.2 | 5.6 | GO:0032982 | myosin filament(GO:0032982) |
| 0.2 | 81.0 | GO:0030141 | secretory granule(GO:0030141) |
| 0.2 | 1.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.2 | 12.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 15.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 12.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 104.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.2 | 4.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 10.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 2.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 14.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 5.1 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 29.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 197.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 7.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 2.4 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 5.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 3.2 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 4.5 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 3.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 7.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 2.3 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 14.7 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.1 | 1.0 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 5.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 4.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 2.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 9.3 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 9.2 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 1.4 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 58.4 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.6 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 2.4 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 6.0 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 1.2 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.4 | 52.3 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 11.4 | 56.9 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 5.7 | 34.0 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 5.2 | 15.6 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 4.3 | 38.5 | GO:0004568 | chitinase activity(GO:0004568) |
| 4.2 | 12.7 | GO:0035375 | zymogen binding(GO:0035375) |
| 3.8 | 18.8 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 3.3 | 19.9 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 3.3 | 13.2 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 3.0 | 12.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 3.0 | 14.8 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 2.8 | 8.5 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 2.8 | 11.1 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 2.5 | 19.9 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 2.3 | 13.5 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 2.2 | 6.7 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 1.9 | 15.3 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 1.9 | 30.4 | GO:0019841 | retinol binding(GO:0019841) |
| 1.8 | 19.3 | GO:0035473 | lipase binding(GO:0035473) |
| 1.7 | 13.6 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 1.7 | 10.0 | GO:1990254 | keratin filament binding(GO:1990254) |
| 1.6 | 9.9 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 1.5 | 4.4 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 1.2 | 14.5 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 1.2 | 17.6 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 1.1 | 10.0 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 1.0 | 15.5 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 1.0 | 8.9 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.9 | 2.6 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.9 | 9.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.8 | 32.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.7 | 13.0 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.7 | 10.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.6 | 4.5 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.6 | 2.6 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.6 | 4.4 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.6 | 17.8 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.6 | 17.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.6 | 3.4 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.6 | 2.2 | GO:0033797 | selenate reductase activity(GO:0033797) |
| 0.5 | 2.2 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.5 | 1.6 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.5 | 3.6 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.5 | 54.6 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.5 | 1.5 | GO:0043404 | corticotrophin-releasing factor receptor activity(GO:0015056) corticotropin-releasing hormone receptor activity(GO:0043404) |
| 0.5 | 3.9 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.5 | 4.8 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.5 | 1.4 | GO:0070002 | glutamic-type peptidase activity(GO:0070002) |
| 0.5 | 18.9 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.5 | 1.4 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.5 | 13.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.5 | 7.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.4 | 4.9 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.4 | 5.2 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.4 | 3.8 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.4 | 1.7 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 0.4 | 10.8 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.4 | 2.9 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.4 | 1.2 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.4 | 10.5 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.4 | 3.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.4 | 9.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.4 | 10.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.4 | 10.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.4 | 1.4 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.3 | 82.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.3 | 1.4 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.3 | 2.9 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.3 | 1.3 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.3 | 42.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.3 | 9.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.3 | 49.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 9.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.3 | 1.8 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 4.7 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.2 | 196.3 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.2 | 40.1 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.2 | 5.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 1.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.2 | 1.5 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.2 | 0.6 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.2 | 2.7 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 2.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.2 | 2.2 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.2 | 3.9 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.2 | 13.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 4.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 1.8 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 5.6 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.2 | 1.4 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 0.1 | 3.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 1.0 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 15.5 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 2.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 9.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 1.7 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 1.7 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.1 | 1.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 3.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 7.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.8 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 7.1 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 3.0 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 10.0 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.1 | 6.6 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.1 | 1.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 1.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 5.0 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.6 | GO:0043199 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) sulfate binding(GO:0043199) |
| 0.1 | 10.0 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.1 | 7.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 2.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.0 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 9.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.8 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.6 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 10.0 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 0.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 3.4 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.1 | 1.0 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.1 | 4.0 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 0.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 1.8 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 6.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 1.8 | GO:0003690 | double-stranded DNA binding(GO:0003690) |
| 0.0 | 1.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 3.2 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 5.0 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 3.4 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 9.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 2.5 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 4.1 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 3.5 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 1.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.4 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 1.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 4.9 | GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors(GO:0016614) |
| 0.0 | 1.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 1.6 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 2.0 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 1.0 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 26.4 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 1.0 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 4.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 3.3 | GO:0003924 | GTPase activity(GO:0003924) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 72.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.8 | 56.0 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.5 | 18.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.5 | 29.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.5 | 33.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.5 | 28.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.4 | 10.8 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.4 | 12.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.4 | 6.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.3 | 88.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.3 | 26.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.3 | 7.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.3 | 11.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.3 | 2.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.3 | 10.7 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.3 | 19.9 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.3 | 23.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.3 | 18.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.2 | 10.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 4.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.2 | 12.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 6.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 5.9 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 1.0 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 3.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 4.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 3.9 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 1.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 12.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 3.9 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 2.8 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 6.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 9.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 1.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 12.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 3.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 4.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.4 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 9.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.6 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 1.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 72.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 1.4 | 15.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 1.2 | 19.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 1.1 | 17.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 1.1 | 13.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.9 | 13.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.9 | 14.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.8 | 24.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.7 | 44.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.5 | 12.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.5 | 1.4 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.5 | 15.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.5 | 16.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.5 | 32.4 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.5 | 10.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.4 | 6.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.4 | 13.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.4 | 3.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.3 | 19.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 5.0 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.3 | 4.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.3 | 6.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.3 | 14.0 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.3 | 9.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.3 | 11.0 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.2 | 14.6 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.2 | 14.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.2 | 1.4 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.2 | 1.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.2 | 3.0 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 10.6 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.2 | 1.4 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 2.8 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 7.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 5.3 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 7.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 2.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 0.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 0.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 1.0 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 2.2 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 1.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 7.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.6 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.0 | 1.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 3.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 3.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.5 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.6 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.8 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 2.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.5 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.4 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |